HER2 also known as ErbB2 is an epidermal growth factor receptor and is a 185 kDa transmembrane glycoprotein. 20–30% of breast cancer patients have overexpression of HER2.1 The extracellular domain of the HER2 with 630 amino acids contains four subdomains I–IV (Figure 1) and studies have revealed that the dimerization loop in domain II helps HER2 to dimerize with other EGFR membrane proteins resulting in cell differentiation and apoptosis.2

Figure 1: Schematic of two different antibodies targeting different regions of the extracellular domain (ECD) of HER2.
The native in-cell receptor is oriented such that the four different binding domains in HER2 are oriented differently with spread out glycosylation sites. According to literature, domains I and II are more accessible than domain IV, which lies tucked very closely to the cell membrane in whole cells.3 In this case, the cell membrane could significantly influence the interaction by imposing a certain amount of steric hindrance to antibodies like Herceptin which target domain IV as seen in Figure 1. Therefore, it is increasingly important to target other accessible domains of HER2 in the native cell environment to understand the true binding kinetics of drug molecules to overcome the clinical problems of drug resistance.4 To effectively do so, it is necessary to explore the epitope space of HER2 for potential clinical targets in a systematic manner.
Surface plasmon resonance microscopy (SPRM) using BI’s SPRm200, integrates label-free, high-resolution SPR binding kinetic measurements with bright-field optical microscopy for the study of biomolecular interactions in the extracellular space of whole cells. It is capable of characterization of binding responses of each cell individually. In this study, the binding kinetics of anti-Her2 antibody targeting the domain II of HER2 of SKBR3 cells was studied using the SPRm200 system. A kinetic titration injection series was performed such that the cells were exposed to serial injections of six anti-HER2 solutions (3.15, 6.25, 12.5, 25, 50, and 100 nM). BI’s Image Analysis software was used for analyzing the SPRM sensorgrams. Regions of activity defined by the red squares in both the bright-field and SPR images produced the binding responses as seen in (Figure 2A- B). A 1:1 kinetic interaction model was fitted to the binding data for kinetics analysis. From the statistical analysis, the kinetic values for the entire cell population were determined to be ka of 9.917 x 105 M-1s-1, kd of 3.058 x 10-3 s, and KD of 2.663 nM (see Figure 2C).
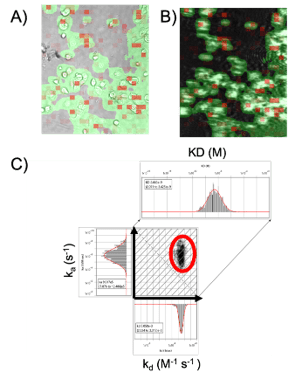
Figure 2: Kinetic interaction analysis was performed on SKBR3 cells using SPRm200: (a) Bright-field image and (b) corresponding SPR image of the SKBR3 cells with red squares representing areas of binding activity (ROIs). C) Every ROI was fit to a 1:1 kinetic binding model to identify areas of binding activity and measure the kinetics of the interaction. Histograms of the kinetic parameters were fitted with Gaussian distributions to extract the mean and 95% confidence interval of the cell population. An affinity isotherm plot is generated from hundreds of responsive regions for anti-HER2 interaction with HER2 receptor indicated using a red circle.
This SPRM study shows that the anti-HER2 targeting domain II of HER2 has a stronger affinity to HER2 than Herceptin, which binds to the evasive domain IV of HER2 in native SKBR3 cells.4 Also, antibodies targeting domain II can effectively block HER2 dimerization which in turn inhibits receptor phosphorylation and disrupts associated cell signaling, proving to be a more effective drug target. A greater understanding of the underlying binding behaviour of HER2-targeting antibodies is crucial to assess their clinical benefits, efficacy, and bio relevance more thoroughly.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published May 20th, 2024
DOWNLOAD PDF
Download a PDF of Application Note 156: Kinetics of Anti-HER2 Binding to Extracellular Domain II of HER2
- Fu, Wenyan, Yuxiao Wang, Yunshan Zhang, Lijuan Xiong, Hiroaki Takeda, Li Ding, Qunfang Xu et al. "Insights into HER2 signaling from step-by-step optimization of anti-HER2 antibodies." In MAbs, vol. 6, no. 4, pp. 978-990. Taylor & Francis, 2014.
- Khoury, T., Mojica, W., Hicks, D., Starostik, P., Ademuyiwa, F., Janarthanan, B., & Cheney, R. T. (2011). ERBB2 juxtamembrane domain (trastuzumab binding site) gene mutation is a rare event in invasive breast cancers overexpressing the ERBB2 gene. Modern Pathology, 24(8), 1055-1059.
- Iqbal, N., & Iqbal, N. (2014). Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Molecular biology international, 2014.
- Dong, Tianbao, et al. "Live cells versus fixated cells: kinetic measurements of biomolecular interactions with the LigandTracer method and surface plasmon resonance microscopy." Molecular Pharmaceutics 20.4 (2023): 2094-2104.
