Understanding antibody binding and kinetics of therapeutic antibodies to tumor cells plays a critical role in advancing therapeutics. SPR is commonly used to study antibody binding reactions. However, it has the following challenges: a) not all antigens can be expressed as soluble proteins, b) techniques used for immobilization of the antigen can affect affinity measurements, and c) purified proteins may not be expressed in the native structure resulting in an altered function.1&2 Hence determining the antibody affinity and avidity on cells, where the antigen is in the native form, becomes critical especially with different emerging antibody modalities.3
SPRM is a label free technology utilized to study binding interactions of membrane proteins without extracting the proteins from the cell, ensuring intact native conformations of the membrane proteins. This technology also enables the study of differences in the affinity and avidity of antibodies with new modalities in the native cell environment.4 In this study, performed in collaboration with Ellaine Mai at Genentech, the effect of avidity and affinity of three different antibodies on MCF-7, a HER2 expressing the cell line, was studied using SPRM. Three structurally different antibodies (Herceptin Ab (bivalent binding to HER2), one-arm anti-HER2 1-Fab IgG (monovalent binding to HER2) and anti-gD, IgG (Isotope control) were used.5
Figure 1: Schematic representation of (A) monovalent binding of one-arm anti-HER2 1-Fab IgG to HER2 and (B) bivalent binding of Herceptin to HER2.
SPRm 200 system combines bright field optical imaging and high-resolution imaging based SPR measurements. SPRM data was analyzed statistically using the provided ImageSPR™ analysis software. Areas that observed biding interactions were marked with colored squares and designated as regions of interest (ROIs) for kinetics analysis. SPRm 200 kinetics analysis of the control anti-gD, IgG exposure to MCF-7 cells revealed negligible on-cell binding interactions, as seen by the sparse number of ROIs in Figure 2.
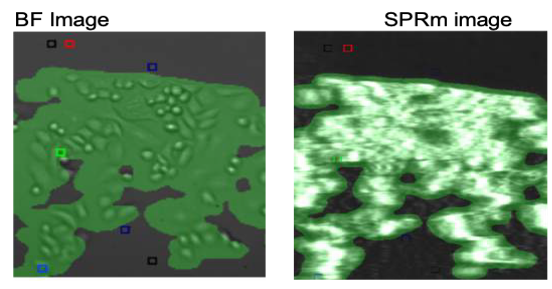
Figure 2: SPRm 200 brightfield image of isotype control (anti-gD,IgG) binding to MCF-7 HER 2 expressing cells; squares indicate ROIs where antibody binding events were detected. Lack of ROI squares in the control shows the lack of binding events.
Significantly more on-cell binding is observed when HER2 binding proteins are exposed to the HER2 expressing MCF-7 cells. As shown in Figure 3, the monovalent anti-HER2 antibody shows only one histogram peak which allows for the determination of one KD. In contrast, the bivalent Herceptin shows two histogram distribution for KD. The KD peak of larger value is attributed to single-arm affinity interactions and corresponds well to the mono-valent KD value. The smaller KD peak with stronger interaction is attributed to avidity effects.
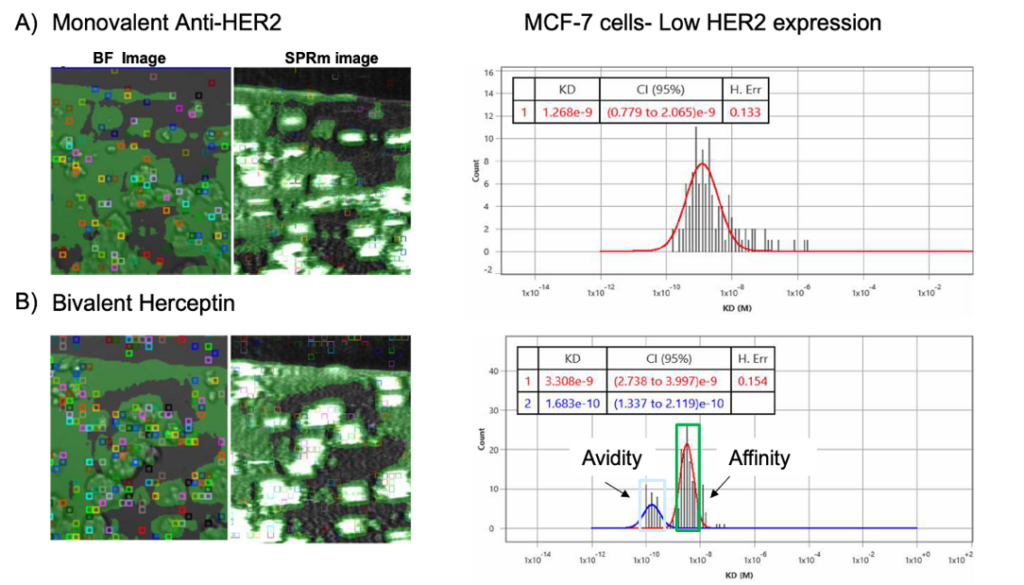
Figure 3: Comparison data of (A) monovalent Anti-Her2 binding to MCF-7 cells and (B) bivalent Herceptin bound to MCF-7 cells; ROI squares on bright field and SPR images indicate areas where antibody binding events were detected. The kinetic binding parameters extracted from every ROI were aggregated to form KD histograms.
| Cell line | Analytes | KD (nM) |
| MCF7 | anti-HER2 IgG (monovalent) | 1.3 |
| MCF7 | Herceptin (bivalent) | 3.3 and 0.17 |
Table1: Kinetic parameters measured from anti-HER2 and Herceptin binding interactions with HER2 in the native in-cell environment of the MCF7 cell line.
Binding interactions with HER2 of the MCF7 cell line show an obvious difference between the monovalent and bivalent forms, highlighting SPRm 200’s unique ability to investigate both affinity and avidity interactions. This capability of SPRm 200 is granted by its powerful sub-cellular resolution and would not be readily possible with the use of lower resolution or end-point measurement approaches.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published Nov 30, 2023
DOWNLOAD PDF
Download a PDF of Application Note 153: Differentiating Mono and Bivalent Antibody Binding in MCF7 Cell line
- Hartl, F. Ulrich, and Manajit Hayer-Hartl. "Converging concepts of protein folding in vitro and in vivo." Nature structural & molecular biology 16.6 (2009): 574-581.
- Das, Anirban, et al. "Rational design of protein-specific folding modifiers." Journal of the American Chemical Society 143.44 (2021): 18766-18776.
- Brinkmann, Ulrich, and Roland E. Kontermann. "The making of bispecific antibodies." In MAbs, vol. 9, no. 2, pp. 182-212. Taylor & Francis, 2017.
- Wang W, Yang Y, Wang S, Nagaraj VJ, Liu Q, Wu J, et al. Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nat Chem. 2012;4(10):846–53. Pmid:23000999.
- Slaga, Dionysos, Diego Ellerman, T. Noelle Lombana, Rajesh Vij, Ji Li, Maria Hristopoulos, Robyn Clark et al. "Avidity-based binding to HER2 results in selective killing of HER2-overexpressing cells by anti-HER2/CD3." Science translational medicine 10, no. 463 (2018): eaat5775.
