Integrins are heterodimeric receptors which help in cell adhesion and are ubiquitous to all metazoans.1 They also play a vital role in numerous cellular processes including cell growth, proliferation, differentiation, migration and phagocytosis of opsonized target.2,3 Integrin, Mac-1 is expressed primarily on myeloid cells such as neutrophils and macrophages. It is involved in the phagocytosis of opsonized particles, cell-cell fusion, and acts as an important regulator of the immune activities of these cells.4 To date, more than 100 ligands have been reported to bind to Mac-1.2
The binding site for the vast majority of ligands is located in the I-domain of Mac-1’s αM subunit (~ 200 residue αMI-domain). Despite the importance of αMI-domain in ligand binding, only the structure of αMI-domain with one of its natural ligands, the complement fragment iC3b, has been determined thus far.5,6 Similar to other integrins, Mac-1 can adopt both inactive and active conformations. The active conformation has an increasing affinity for its ligands; most studies of αMI-domain’s interactions with ligands use active αMI-domain.7 Ligand specificities of integrins are vital determinants of their activities, and understanding the ligand binding mechanisms behind these specificities is critical to develop drugs targeting integrins.
Figure 1: Schematic of SPR measurement of Mac-1 receptor αMI-domain.
In this study, three mutants of αMI-domain were purified and characterized using SPR8 as seen in Figure 1. Among the three mutants (ΔK315, I316G, and Q163C/Q309C) only one of the mutants (Q163C/Q309C) can spontaneously form the required disulfide bond to maintain the active conformation of the αMI-domain and has a better yield with thermal stability. Studies claim that HEK293T cells expressing Mac-1 with the Q163C/Q309C mutations were shown to have a higher affinity for ligands than wild type Mac-1 expressing cells, but there is no exclusive biochemical study showing the Q163C/Q309C αMI-domain has enhanced affinity for ligands compared to wild type I-domain.9 Surface Plasmon Resonance (SPR) was used to understand the affinity of the isolated mutants with C3d and to investigate the enhanced affinity of Q163C/Q309C mutants.
SPR analysis was carried out on a BI-4500A SPR instrument (Biosensing Instrument Inc.). Figure 2 shows the SPR sensorgrams for the αMI-domain variants flowed over a sensor with immobilized C3d. These sensorgrams fitted to a one-to-one binding model showed that, Q163C/Q309C, ΔK315, and I316G mutants all have dissociation constant of binding (Kd) similar to other reported studies. The results indicated that the Q163C/Q309C mutant has a similar affinity for C3d as other active mutants. However, the koff rate of ΔK315 appears to be slower than the other two active mutants, implying the kinetics of binding differ slightly among the mutants. Control experiments carried out using inactive αMI-domain showed inactive αMI-domain does not have a measurable affinity for C3d.
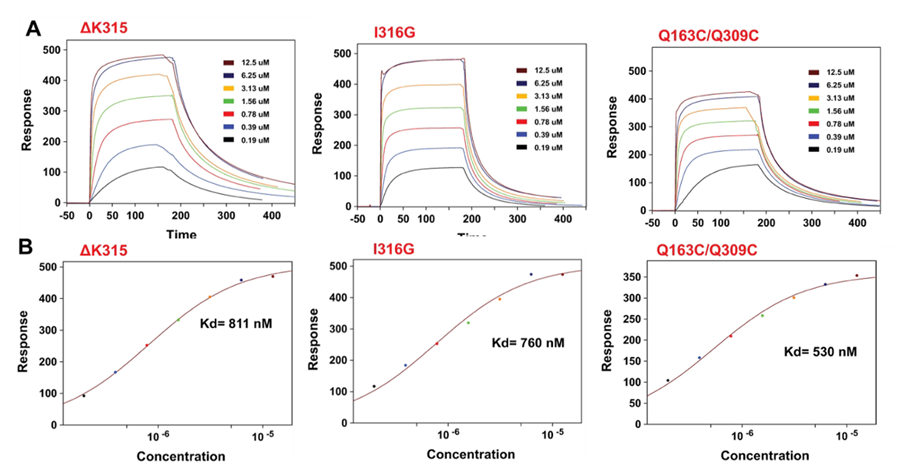
Figure 2: SPR characterization of C3d’s interactions with active αMI-domains. Before measurement, 50 μM C3d was flowed at a rate of 20 μL/min over EDC/NHS-activated CM-dextran sensor until a response of ~ 1000 RU was observed. An increase of 1 RU corresponds to protein deposition of ~ 1 pg / mm2. A) Sensorgrams of a C3d-functionalized sensor treated with 0.19, 0.39 0.78, 1.56, 3.13, 6.25, and 12.5 μM of ΔK315, I316G, or Q163C/Q309C mutants. The sensor was regenerated with 1.5 M NaCl after each sample. Injection of αMI-domain started at time zero and ended after 180 seconds. B) Estimation of the dissociation constant (kd) of the interaction for each active αMI-domain by fitting the equilibrium values of the response curves to a one-to-one binding model. Response curves were background corrected by subtracting the response of the reference channel with no immobilized C3d.
The integrin Mac-1 is an important integrin involved in many aspects of leukocyte biology. Understanding the mechanisms of its ligand specificity is essential to developing targeted treatments against Mac-1. However, the lack of a suitable active αMI-domain has so far prevented investigating the interactions of active αMI-domain with its ligands using BI-4500A SPR instrument. This report systematically examined known active mutants of αMI-domain and showed that the Q163C/Q309C mutant adopts the active conformation and has an enhanced affinity for ligands compared to wild type I-domain. The availability of such a mutant will enable more SPR studies of αMI-domain’s interaction with ligands and reveal more insights into the mechanisms of Mac-1 activity.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published October 30, 2023
DOWNLOAD PDF
Download a PDF of Application Note 152: Binding Studies of an Integrin and its Mutants using BI-4500
- Luo BH, Carman CV, Springer TA (2007) Structural basis of integrin regulation and signaling. Annu Rev Immunol 25: 619–647. https://doi.org/10.1146/annurev.immunol.25.022106.141618 PMID: 17201681
- Lamers C, Pluss CJ, Ricklin D (2021) The Promiscuous Profile of Complement Receptor 3 in Ligand Binding, Immune Modulation, and Pathophysiology. Front Immunol 12: 662164. https://doi.org/10. 3389/fimmu.2021.662164 PMID: 33995387
- Vorup-Jensen T, Jensen RK (2018) Structural Immunology of Complement Receptors 3 and 4. Front Immunol 9: 2716. https://doi.org/10.3389/fimmu.2018.02716 PMID: 30534123
- Rosetti F, Mayadas TN (2016) The many faces of Mac-1 in autoimmune disease. Immunol Rev 269: 175–193. https://doi.org/10.1111/imr.12373 PMID: 26683153
- Bajic G, Yatime L, Sim RB, Vorup-Jensen T, Andersen GR (2013) Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor Proc Natl Acad Sci USA 110: 16426–16431. https://doi.org/10.1073/pnas.1311261110 PMID: 24065820
- Fernandez FJ, Santos-Lopez J, Martinez-Barricarte R, Querol-Garcia J, Martin-Merinero H, et al. (2022) The crystal structure of iC3b-CR3 alphaI reveals a modular recognition of the main opsonin iC3b by the CR3 integrin receptor. Nat Commun 13: 1955.
- Lee JO, Bankston LA, Arnaout MA, Liddington RC (1995) Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure 3: 1333–1340. https://doi.org/10.1016/s0969-2126(01) 00271-4 PMID: 8747460
- Nguyen, Hoa, Tianwei Jing, and Xu Wang. "The Q163C/Q309C mutant of αMI-domain is an active variant suitable for NMR characterization." Plos one 18.1 (2023): e0280778.
- Shimaoka M, Lu C, Salas A, Xiao T, Takagi J, et al. (2002) Stabilizing the integrin αM inserted domain in alternative conformations with a range of engineered disulfide bonds. Proceedings of the National Academy of Sciences 99: 16737–16741.
