Inflammatory bowel disease (IBD) is a complex, multifactorial inflammatory disease of the gastrointestinal tract resulting in chronic, aberrant mucosal inflammation of the gastrointestinal tract.1,2 The combination of immunologic, environmental, microbiome, and genetic factors is believed to contribute to disease onset and continued progression. In active ulcerative colitis (UC) and Crohn’s disease (CD), the intraluminal colonic pH is significantly lower than normal individuals. It has been suggested that pH sensing receptors could contribute to the onset and progression of diseases such as IBD.3
The proton-activated G protein–coupled receptors (GPCRs) like GPR4, TDAG8 (or GPR65), and OGR1 (or GPR68) are pH sensors that can trigger intracellular signaling in response to alterations in extracellular pH around physiological values, i.e., in the range between pH 7.5 and 6.5.4 GPR4 is predominantly expressed in endothelial cells (ECs) and GPR4 activation has been shown to increase the expression of numerous inflammatory and stress response genes in vascular ECs, promoting leukocyte infiltration in the mucosa.5 Absence of GPR4 has been shown to ameliorate colitis in several murine IBD models. Therefore, GPR4 inhibition has been proposed as a potential beneficial therapeutic approach for the treatment of IBD.
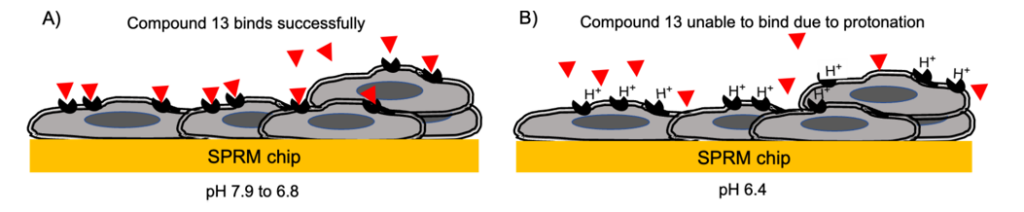
Figure 1: Schematic of A) Compound 13 (red triangles) binding to the black GPR4 receptors on the cell surface at pH7.9 to 6.8 and B) the inhibition of binding at pH 6.4 due to the protonated state of the GPR4 receptors.
To explore the therapeutic potential of GPR4 antagonism in IBD, Compound 13, a reported selective GPR4 antagonist, was tested in this study. Compound 13 has been described as a selective, orally bioavailable GPR4 antagonist with a favorable safety and pharmacokinetic profile.6 Despite reasonable exposures consistent with previous reports, Compound 13 was ineffective at preventing colitis in IL10-/- mice.7 The lack of apparent and significant target engagement led to investigating the mechanism of inhibition of Compound 13. Literature suggests that the performance Compound 13 highly depends on pH as it behaves as an orthosteric antagonist, i.e., loss of potency at low pH (high concentration of agonist present).8 However, the concept of orthosteric antagonism for pH sensing receptors remains poorly characterized.
Surface plasmon resonance microscopy (SPRM) measurements were used to confirm the binding capabilities of Compound 13 at different pH conditions. SPRM experiments were carried out on the SPRm 200AP system, which combines bright field optical imaging and high-resolution imaging-based SPR measurement. SPR has been the go-to technique for measuring binding affinity and kinetics in a label-free manner. However, measuring membrane protein binding with SPR has been a challenge because of the need to immobilize purified receptors on the sensor chip surface. SPRM is a label free technology utilized to study binding interactions of membrane proteins without extracting the proteins from the cell, ensuring intact native conformations of the membrane proteins.9 SPRM data was analyzed statistically using the provided ImageSPR™ analysis software. The measured SPRM images were divided into about 600 small regions of interest (ROIs), about 20×20 µm each. SPR sensorgrams were generated for each ROI for all concentrations injected.
SPRM 200 analysis in hGPR4-expressing cells revealed that Compound 13 could bind to hGPR4 at both pH 7.9 and 6.8 with equilibrium dissociation constants of 3.9 and 5.5 nM respectively (Figure 2). No binding was detected at pH 6.4.10 No binding was detected either at pH 7.9 and pH 6.8 in mock cells that did not express hGPR4. Also, Mutagenesis studies confirmed Compound 13 likely binds to the conserved orthosteric binding site in G protein-coupled receptors, where a histidine sits in GPR4 likely preventing Compound 13 binding when protonated in acidic conditions.11 It has been proposed that multiple residues in GPR4 are protonated in low pH conditions, resulting in conformation changes which could also prevent Compound 13 from binding to GPR.
SPRm 200 measurements largely confirmed Compound 13 preferentially binds to the inactive conformation of GPR4. An interesting advantage of SPRm is its ability to measure binding events to multiple regions of the cells, capturing the diversity in receptor conformations.
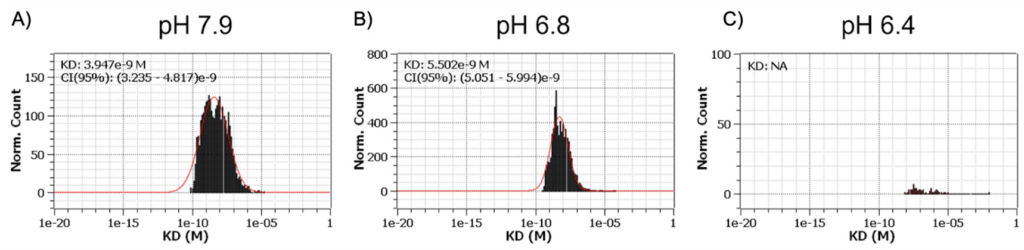
Figure 2: Compound 13 only binds to GPR4 in its inactive conformation. Binding affinities of Compound 13 to human GPR4 were assessed by SPRm 200 at various pHs. To lock GPR4 in either its active or inactive conformation, human GPR4-expressing cells were stimulated by (A) pH 7.9, (B) pH 6.8, or (C) pH 6.4 – adjusted buffers and then fixed right away before flowing Compound 13 (in pH 7.4 buffer) for binding determination. The measured SPRM images were divided into about 600 small regions of interest (ROIs), about 20×20 μm each. SPRM sensorgrams were generated for each ROI for all concentrations injected. The histograms represent the distribution of measured thermodynamic KD from all isothermal curves.
A number of high affinity subnanomolar binding events at pH 7.9 were observed, that were absent at pH 6.8 and 6.4, consistent with the affinity of Compound 13 decreasing at lower pH (Figure 3). A small number of binding events were detected at pH 6.4, but they were not sufficient to generate a statistically significant histogram distribution. Thus, SPRM measurements largely confirmed Compound 13 preferentially binds to the inactive conformation of GPR4. Thermodynamic analysis was consistent with what was previously observed in that Compound 13 could not bind to GPR4 at pH 6.4.
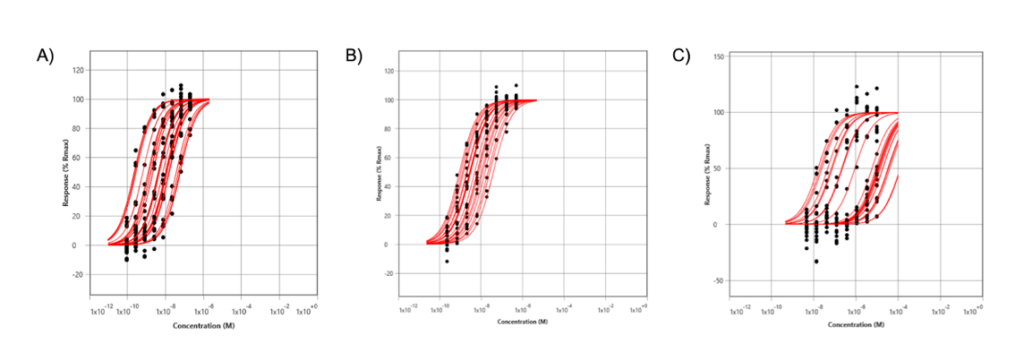
Figure 3: SPRm200 isothermal curves of Compound 13 binding to multiple regions of hGPR4-expressing HeLa cells, at different pHs: (A) pH 7.9, (B) pH 6.8, and (C) pH 6.4.
Compound 13 binds to the conserved GPCRs orthosteric binding site, so it behaves as an orthosteric antagonist, only binding to GPR4 in its inactive, unprotonated state. As the degree of inflammation is generally correlated with increased acidity, Compound 13 is unlikely the appropriate compound to study GPR4 and its role in inflammatory bowel conditions. Compound 13 and its analogues’ pH dependence strongly limits their utility in studying the role of GPR4 in acidosis-associated moderate-to-severe inflammatory conditions. While there is no published precedent for proton-sensing receptors, the thermodynamic study of the role of antagonists for pH-sensing GPCRs using SPRM demonstrates here to be a useful step in drug discovery
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published August 1, 2023
DOWNLOAD PDF
Download a PDF of Application Note 150: Studying the role of an antagonist for a pH-sensing GPCR using SPRm 200
- Andersen V, Ernst A, Christensen J, Ostergaard M, Jacobsen BA, Tjonneland A, Krarup HB and Vogel U (2010) The polymorphism rs3024505 proximal to IL-10 is associated with risk of ulcerative colitis and Crohn’s disease in a Danish case-control study. BMC Med Genet 11:82.
- Cummins EP and Crean D (2017) Hypoxia and inflammatory bowel disease. Microbes Infect 19:210-221.
- Anderson M, Moshnikova A, Engelman DM, Reshetnyak YK and Andreev OA (2016) Probe for the measurement of cell surface pH in vivo and ex vivo. Proc Natl Acad Sci U S A 113:8177-8181.
- Imenez Silva PH and Wagner CA (2022) Physiological relevance of proton-activated GPCRs. Pflugers Arch.
- Dong L, Li Z, Leffler NR, Asch AS, Chi JT and Yang LV (2013) Acidosis activation of the proton-sensing GPR4 receptor stimulates vascular endothelial cell inflammatory responses revealed by transcriptome analysis. PLoS One 8:e61991.
- Sanderlin EJ, Marie M, Velcicky J, Loetscher P and Yang LV (2019) Pharmacological inhibition of GPR4 remediates intestinal inflammation in a mouse colitis model. Eur J Pharmacol 852:218-230.
- Rennick DM and Fort MM (2000) Lessons from genetically engineered animal models. XII. IL10-deficient (IL-10(-/-) mice and intestinal inflammation. Am J Physiol Gastrointest Liver Physiol 278:G829-833.
- Tobo A, Tobo M, Nakakura T, Ebara M, Tomura H, Mogi C, Im DS, Murata N, Kuwabara A, Ito S, Fukuda H, Arisawa M, Shuto S, Nakaya M, Kurose H, Sato K and Okajima F (2015)
- Wang W, Yang Y, Wang S, Nagaraj VJ, Liu Q, Wu J and Tao N (2012) Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nat Chem 4:846-853.
- Stalewski, Jacek, Amy Y. Shih, Romeo Papazyan, Jocelyn Ramirez, Gerardo Ibanez, Peng Hsiao, Yong Yue et al. "pH dependence of a GPR4 selective antagonist hampers its therapeutic potential." Journal of Pharmacology and Experimental Therapeutics (2023).
- Rowe JB, Kapolka NJ, Taghon GJ, Morgan WM and Isom DG (2021) The evolution and mechanism of GPCR proton sensing. J Biol Chem 296:100167.
