Membrane proteins play a major role in cell biology and are an important class of receptors targeted by drug candidates being developed in the pharmaceutical industry.1,2 Techniques for studying the kinetic interaction of drug ligands with target protein receptors in their native cell-based form has proven critical in the drug development process.3-5 One such powerful cell-based technology for studying binding interaction kinetics, Surface Plasmon Resonance Microscopy (SPRM), does not require labeled ligands and is amenable to the screening of antibody candidates and small molecules alike.6,7 Furthermore, the high-resolution capability of SPRM enables subcellular analysis which facilitates observation of heterogeneous behavior.
With SPRM, cells are typically attached to a biopolymer coated (e.g., polylysine, fibronectin, and collagen8,9) gold substrate. Both live or fixed cells may be studied with SPRM, but in most cases the latter results in longer, more stable experiments. Fixation occurs by exposing cells to a crosslinking reagent, e.g. paraformaldehyde (PFA).10-12 PFA stabilizes the cell by cross-linking proteins and thereby maintaining the structure of epitopes (Figure1). However, at times fixation may block epitopes hidden within the folded protein. Alternatively, fixation can be performeded by using organic solvents like methanol. This is beneficial for when cells/receptors are sensitive to crosslinking agents, or for revealing linear epitopes previously hidden within the protein.13 However, this approach can be too harsh, causing dehydration and precipitation of cellular components, and possibly even reducing the activity of certain membrane proteins.
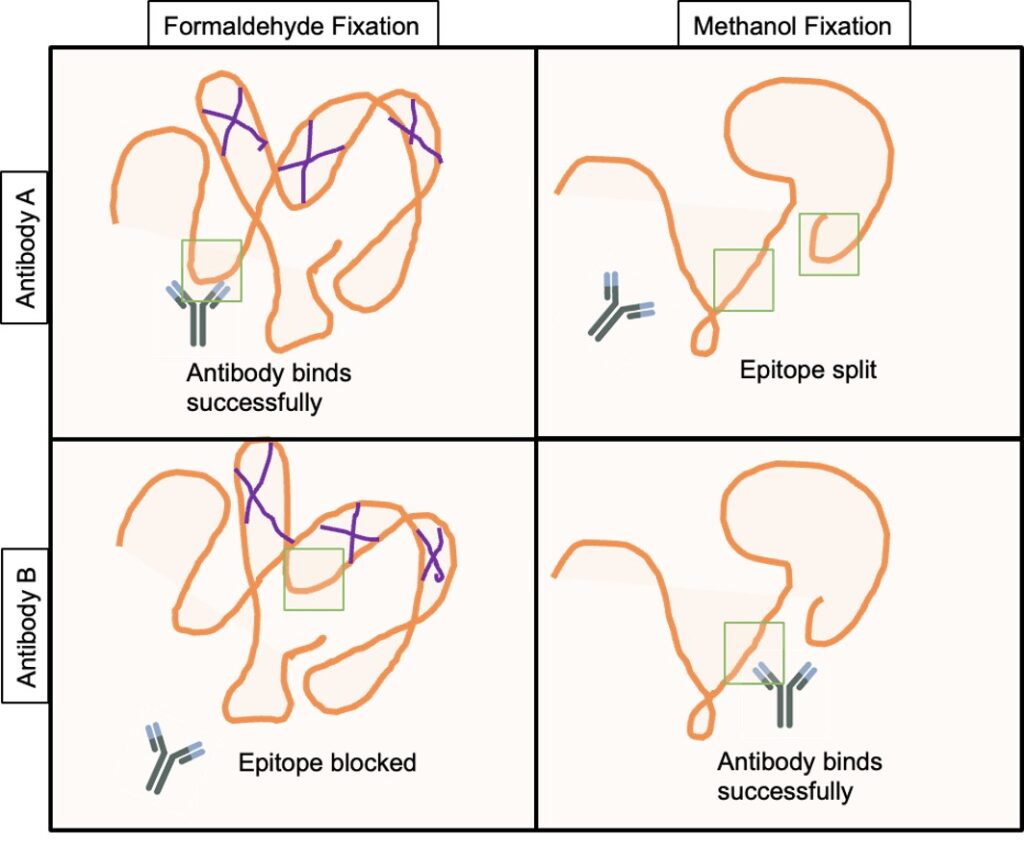
Figure 1: Schematic of the impacts of fixation methods on antibody binding. PFA fixation cross-links proteins (purple lines), which maintains structural epitopes (green squares), but can also block epitopes hidden within the protein. Methanol fixation avoids crosslinking, but dehydrates proteins, which can disrupt protein structure but also expose epitopes previously hidden within the protein.
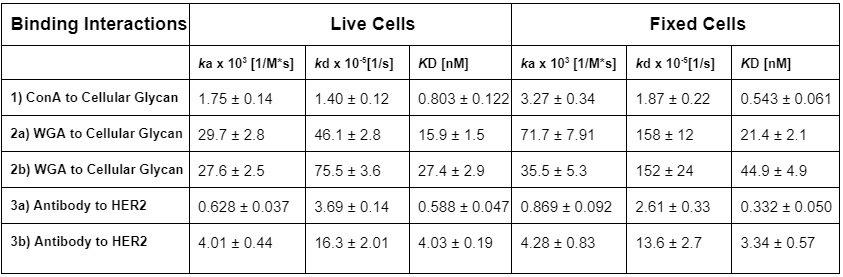
Table 1: Kinetic interaction results with and without fixation
To better understand the effects of fixation on receptor-ligand binding affinity and kinetics, Tiambao et al. performed the first of its kind experiment whereby three biomolecular interactions were examined on both live and fixed cells under the same experimental conditions.14 In this study, the following interactions were studied, (1) concanavalin A (ConA) binding to α-D-mannosyl glycan on the SKBR3 breast cancer cells, a 1:1 binding reaction; (2) attachment of wheat germ agglutinin (WGA) to the N-acetyl glucosamine (GlcNAc) and N-acetyl-neuraminic acid (NeuNAc) types of glycans on HFF cells, a 1:2 binding reaction; and (3) the specific binding of an antibody to human epidermal growth factor receptor 2 (HER2) receptors on the SKBR3 cells, a 1:2 binding reaction.
In general, it was found that cell fixation tends to slightly accelerate the ligand association rate while also slightly decelerate the ligand dissociation rate, keeping the affinity value nearly unchanged. On SKBR3 cells, the slight effect of fixation on binding kinetics is explained by a less mobile HER2 receptor being confined in a local environment of partially interconnected protein molecules. However, the slight differences observed in the binding kinetics between live and fixed cells are considered well within the typical error range attributed to cellular heterogeneity and measurement error. It is worth noting that PFA is reported to only link 3−22% amino acid residues of certain membrane proteins.15 Tanaka et al. explains that though fixation leads to cell shrinkage and a more stiffened membrane, many membrane proteins can remain mobile after fixation.14-18 For example, in the membrane of fixed human T24 cells, a large fraction of proteins can still diffuse laterally.19
Consequently, in the three cases studied, the orientation and overall structure of the receptors do not appear to be affected significantly by chemical fixation. This contention is supported by the fact that the binding affinities of the three ligands in this specific study are comparable between fixed and live cells. Tianbao et al. shows that for cell constituents whose structures and functions are not closely dependent on cell viability, the ligand binding kinetics of fixed cells is only slightly different from that of live cells.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published July 18, 2023
DOWNLOAD PDF
Download a PDF of Application Note 149: The Effect of Cell Fixation on Binding Interaction Kinetics
- Fagerberg, L.; Jonasson, K.; von Heijne, G.; Uhlen, M.; Berglund, L., Prediction of the Human Membrane Proteome. Proteomics 2010, 10 (6), 1141-1149.
- Gulezian, E.; Crivello, C.; Bednenko, J.; Zafra, C.; Zhang, Y.; Colussi, P.; Hussain, S., Membrane Protein Production and Formulation for Drug Discovery. Trends Pharmacol. Sci. 2021, 42 (8), 657-674
- Michelini, E.; Cevenini, L.; Mezzanotte, L.; Coppa, A.; Roda, A. Cell-Based Assays: Fuelling Drug Discovery. Anal. Bioanal. Chem. 2010, 398, 227−238.
- Xi, B.; Yu, N.; Wang, X.; Xu, X.; Abassi, Y. A. The Application of Cell-Based Label-Free Technology in Drug Discovery. Biotechnol. J. 2008, 3, 484−495.
- Chen, J. Y.; Penn, L. S.; Xi, J., Quartz Crystal Microbalance: Sensing Cell-Substrate Adhesion and Beyond. Biosens. Bioelectron. 2018, 99, 593-602.
- Wang, W.; Yang, Y.; Wang, S.; Nagaraj, V. J.; Liu, Q.; Wu, J.; Tao, N., Label-Free Measuring and Mapping of Binding Kinetics of Membrane Proteins in Single Living Cells. Nat. Chem. 2012, 4 (10), 846-853.
- Zhou, X.-L.; Yang, Y.; Wang, S.; Liu, X.-W., Surface Plasmon Resonance Microscopy: From Single-Molecule Sensing to Single-Cell Imaging. Angew. Chem. Int. Ed. Engl. 2020, 59 (5), 1776-1785.
- Li, X.; Yao, J.; Yang, X.; Tian, W.; Liu, L., Surface Modification with Fibronectin or Collagen to Improve the Cell Adhesion. Appl. Surf. Sci. 2008, 255 (2), 459-461.
- Mazia, D.; Schatten, G.; Sale, W., Adhesion of Cells to Surfaces Coated with Polylysine. Applications to Electron Microscopy. J. Cell Biol. 1975, 66 (1), 198-200.
- Fox, C. H.; Johnson, F. B.; Whiting, J.; Roller, P. P., Formaldehyde Fixation. J. Histochem. Cytochem. 1985, 33 (8), 845-853.
- Tanaka, K. A. K.; Suzuki, K. G. N.; Shirai, Y. M.; Shibutani, S. T.; Miyahara, M. S. H.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T. K.; Kusumi, A., Membrane Molecules Mobile Even after Chemical Fixation. Nat. Methods 2010, 7 (11), 865-866.
- Kim, S.-O.; Kim, J.; Okajima, T.; Cho, N.-J., Mechanical Properties of ParaformaldehydeTreated Individual Cells Investigated by Atomic Force Microscopy and Scanning Ion Conductance Microscopy. Nano Converg. 2017, 4 (1), 5.
- Bull, H. B.; Breese, K. R. Interaction of alcohols with proteins. Biopolymers 1978, 17, 2121−2131.
- Dong, Tianbao, et al. "Live Cells versus Fixated Cells: Kinetic Measurements of Biomolecular Interactions with the LigandTracer Method and Surface Plasmon Resonance Microscopy." Molecular Pharmaceutics (2023).
- Metz, B.; Kersten, G. F. A.; Hoogerhout, P.; Brugghe, H. F.; Timmermans, H. A. M.; de Jong, A.; Meiring, H.; ten Hove, J.; Hennink, W. E.; Crommelin, D. J. A.; Jiskoot, W. Identification of Formaldehyde-induced Modifications in Proteins. J. Biol. Chem. 2004, 279, 6235−6243.
- Tanaka, K. A. K.; Suzuki, K. G. N.; Shirai, Y. M.; Shibutani, S. T.; Miyahara, M. S. H.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T. K.; Kusumi, A., Membrane Molecules Mobile Even after Chemical Fixation. Nat. Methods 2010, 7 (11), 865-866.
- Kim, S.-O.; Kim, J.; Okajima, T.; Cho, N.-J., Mechanical Properties of ParaformaldehydeTreated Individual Cells Investigated by Atomic Force Microscopy and Scanning Ion Conductance Microscopy. Nano Converg. 2017, 4 (1), 5.
- Iqbal, N.; Iqbal, N., Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol. Biol. Int. 2014, 2014, 852748.
- Tanaka, K. A. K.; Suzuki, K. G. N.; Shirai, Y. M.; Shibutani, S. T.; Miyahara, M. S. H.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T. K.; Kusumi, A. Membrane Molecules Mobile Even after Chemical Fixation. Nat. Methods 2010, 7, 865−866.
