Chemoattractant receptors and their cognate ligands are promising drug targets for disruption of the tumor progression cascade. Most of these chemoattractant receptors are conventional seven transmembrane-spanning (7TM) G-coupled protein receptors (GPCRs)1,2. ACKRs ( Atypical chemokine receptors), unlike conventional GPCRs, do not activate the G-protein dependent signaling required to direct cell migration3,4. Rather, they modulate chemokine availability by acting as scavenger receptors, promoting chemokine endocytosis, or creating chemokine gradients.
C-C motif chemokine receptor-like 2 (CCRL2) is a non-signaling 7TM receptor related to the ACKR family5. However, CCRL2 differs from ACKRs because it is devoid of ligand scavenger functions and lacks confirmed binding to classical chemokines. CCRL2 is a non-signaling receptor that binds chemotactic ligands to shape leukocyte recruitment to sites of inflammation. However, there is a lack of knowledge on the functional impact and the types of ligands that directly bind CCRL2.
Recent technological advancements in the field of surface plasmon resonance has facilitated the characterization of ligand-receptor interactions under native expression conditions. Surface Plasmon Resonance (SPR) Microscopy incorporates SPR and brightfield microscopy (Fig 1A) to detect binding of unlabeled molecules to cell surface targets.6-8
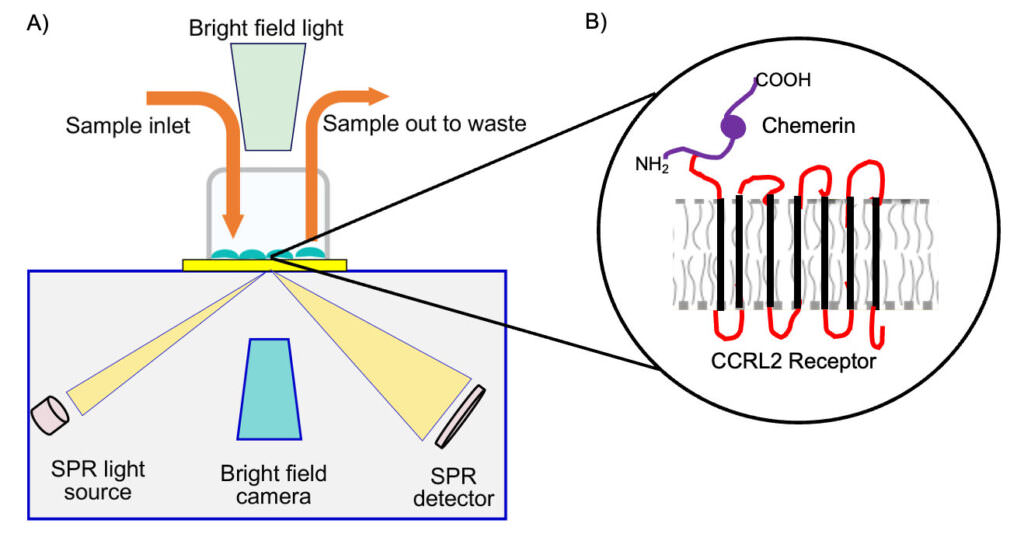
Figure 1: Schematic representation of (A) SPRM instrument depicting the SPR and brightfield input, cell chamber and sample flow.9 (B) Chemerin binding to transmembrane CCRL2 receptor.
SPRM measures the kinetics of molecular interactions to the surface of whole cells and has been crucial for determining on-rate (ka), off-rate (kd) and binding affinity (KD) for ligands to cell-surface expressed receptors. Since SPRM does not require tagged or labeled ligands, it is beneficial for mitigating the potential adverse effects of labeling on ligand function and affinity. Using BI’s ImageSPR™analysis software, the SPRM sensor surface is uniformly divided into hundreds of regions of interest (ROIs). ROIs that detect a binding response are fit to kinetic interaction models for statistical analysis.
In this study, a BI SPRm200 instrument was used to confirm that chemerin, a non-chemokine chemotactic protein ( which was not reported previously) binds to CCRL2 (Fig 1B) on parental HEK 293T cells as well as the variant HEKhuCCRL2 cells9 . Numerous ROIs were observed over cells when chemerin was exposed to HEK-huCCRL2 cells (Fig 2A), but only a few ROIs were present when chemerin was exposed toh parental HEK cells (Fig 2B), indicating that chemerin specifically binds cells expressing CCRL2. Kinetic data assessing chemerin binding to HEK-huCCRL2 cells were fitted to 1:1 binding model show robust gaussian distributions for calculated on-rate, off-rate and binding affinity (Fig 2 C, D and E), whereas minimal binding of chemerin to parental HEK cells was detected and KD values could not be calculated. The mean values for chemerin binding HEK-huCCRL2 cells across all experiments performed for on-rate (ka = 3.11E+05 M-1 s -1), off-rate (kd = 1.16E-03 s -1) and binding affinity (KD = 5.49 nM) were reported. Importantly, the mean KD of chemerin binding to huCCRL2 determined by SPRM was comparable to previously reported binding affinity (2.35 nM) determined by radiolabeled chemerin binding to cells overexpressing huCCRL210.
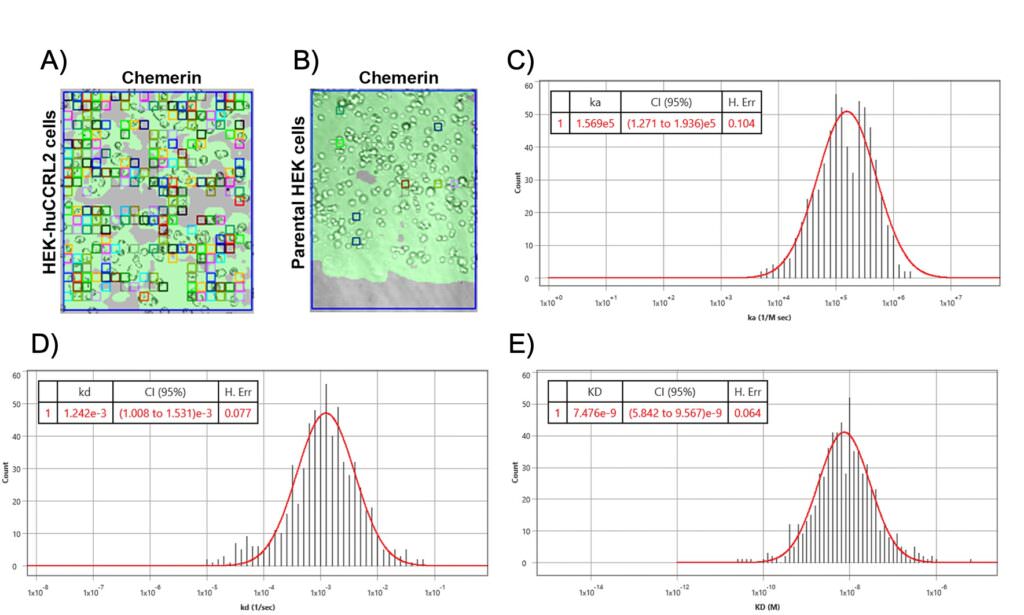
Figure 2: SPRm200 brightfield image of (A) human chemerin bound to HEK-huCCRL2 cells or (B) human chemerin bound to parental HEK cells; squares indicate ROIs where chemerin binding events were detected and are representative of N = 3. Representative histogram distributions for (C) on-rate (ka), (D) off-rate (kd) and (E) binding affinity (KD) were generated from kinetic fit of ROI sensorgrams for human chemerin binding to HEK-huCCRL2 cells and are representative experiments of N = 3. Histograms of on-rate (C) and off-rate (D) values generated using Biosensing Instrument Image Analysis Software are assigned units of ka (1/M sec) and kd (1/sec), respectively.
To investigate whether biotin-labeling may interfere with ligand binding to HEK-huCCLR2 cells, a subset of label-free C-C chemokines was assessed by an SPRm200 instrument. Few ROIs were present when a subset of label-free chemokines like CCL2 and CCL5 were incubated with HEK-huCCRL2 cells and KD values could not be calculated. However the kinetic values of biotin-chemerin binding to HEK-huCCRL2 cells were consistent with the label-free chemerin values as seen in Table 1.
Biotinylation can occur at any reactive primary amine, and an SPRM assessment of the chemerin binding to HEKhuCCRL2 cells shows negligible effects on binding due to biotinylation.
| Chemerin and CCRL2 Interaction | ka x 105 [1/M*s] | kd x 10-3[1/s] | KD [nM] |
| Unlabeled chemerin | 3.11 | 1.16 | 5.49 |
| Biotinylated chemerin | 2.97 | 1.84 | 8.55 |
Table 1: Summary of chemerin binding to HEK-huCCRL2
SPRM enabled direct binding and kinetics analysis of a previously unreported C-C chemokine. The binding kinetics of Chemerin to CCRL2 on parental HEK 293T cells as well as the variant HEKhuCCRL2 cells were determined.9
CCRL2 does not activate intracellular G proteins and concentrates its ligand at the plasma membrane11. The downstream signaling of this receptor is not known and identifying tools that neutralize CCRL2 from ligand binding is the main way to identify its function. In order to do so, SPRM was used as a confirmatory antibody binding method to study the binding kinetics of two anti-huCCRL2 antibody clones (K097F7 and 152254) to HEK-huCCRL2 cells and parental HEK 293T cells. Numerous ROIs were detected on HEK- huCCRL2 cells (Fig 3A and 3B), but not on parental HEK cells.
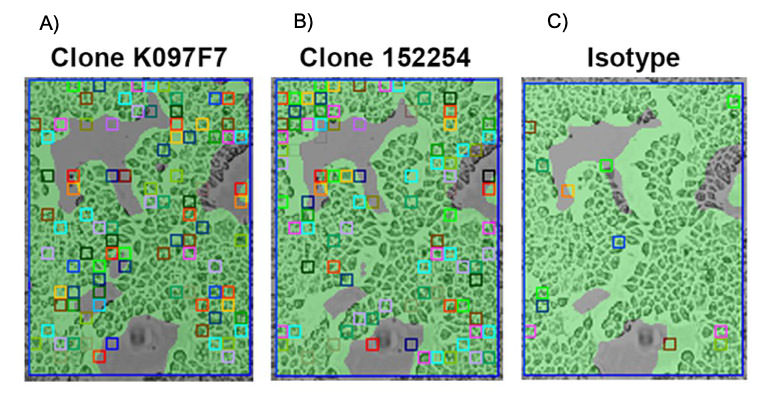
Figure 3: SPRm200 brightfield image of (A) K097F7 binding to HEK-huCCRL2 cells, (B) 152254 binding to HEK-huCCRL2 cells, and (C) isotype control binding to HEK- huCCRL2 cells; squares indicate ROIs where antibody binding events were detected.
| Neutralizing Ab and CCRL2 Interaction | ka x 105 [1/M*s] | kd x 10-3[1/s] | KD [nM] |
| K097F7 | 5.65 | 8.28 | 2.48 |
| 152254 | 5.88 | 1.94 | 4.75 |
Table 2 : Summary of neutralizing antibody clones binding to HEK-huCCRL2
The binding affinity (KD) for K097F7 and 152254 to HEK-huCCRL2 cells was 2.48 nM and 4.75 nM, respectively (Table 2). Importantly, minimal binding of K097F7 and 152254 to parental HEK cells resulted in KD values that could not be calculated. Moreover, minimal binding of an isotype control to HEK-huCCRL2 cells was observed (Fig 3C), indicating that K097F7 and 152254 bound specifically to CCRL2.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published July 17, 2023
DOWNLOAD PDF
Download a PDF of Application Note 148: New Discovery of Chemerin Binding to CCRL2 Receptor Using SPRm 200
- Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12(2):121–7. Pmid:10714678
- Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36(5):705–16. Pmid:22633458
- Nibbs RJB, Graham GJ. Immune regulation by atypical chemokine receptors. Nat Rev Immunol. 2013;13(11):815–29. Pmid:24319779
- Bonecchi R, Graham GJ. Atypical chemokine receptors and their roles in the resolution of the inflammatory response. Front Immunol. 2016;7:224. Pmid:27375622
- Schioppa T, Sozio F, Barbazza I, Scutera S, Bosisio D, Sozzani S, et al. Molecular basis for CCRL2 regulation of leukocyte migration. Front Cell Dev Biol. 2020;8:615031. Pmid:33363177
- Wang W, Yang Y, Wang S, Nagaraj VJ, Liu Q, Wu J, et al. Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nat Chem. 2012;4(10):846–53. Pmid:23000999
- Wang W, Yin L, Gonzalez-Malerva L, Wang S, Yu X, Eaton S, et al. In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance. Sci Rep. 2014;4:6609. Pmid:25312029
- Zhang F, Wang S, Yin L, Yang Y, Guan Y, Wang W, et al. Quantification of epidermal growth factor receptor expression level and binding kinetics on cell surfaces by surface plasmon resonance imaging. Anal Chem. 2015;87(19):9960–5. Pmid:2636833
- Su, Z., Brooks, J., Pelker, J., Andreyeva, T., Sobon, H., Gifford, R., Powers, M., Wang, J., Dower, C., Hegen, M. and Messing, D., 2023. Studies with neutralizing antibodies suggest CXCL8-mediated neutrophil activation is independent of CC motif chemokine receptor-like 2 (CCRL2) ligand binding function. PloS one, 18(1), p.e0280590.
- De Henau O, Degroot GN, Imbault V, Robert V, De Poorter C, McHeik S, et al. Signaling properties of chemerin receptors CMKLR1, GPR1 and CCRL2. PLOS One. 2016;11(10):e0164179. pmid:27716822.
- Del Prete A, Bonecchi R, Vecchi A, Mantovani A, Sozzani S. CCRL2, a fringe member of the atypical chemoattractant receptor family. Eur J Immunol. 2013;43(6):1418–22. pmid:23580473
