SPRM technology is unique in its ability to measure label-free kinetic binding interactions on cells in real-time. Its powerful single-cell resolution enables detailed study of cellular heterogeneity as well as off-target behaviors such as nonspecific interactions. For this reason, nonspecific binding interactions may be more readily identified and removed, resulting in a more accurate kinetic interaction analysis.
SPRM experiments done with SPRm200 consist of a layer of cells bound to the sensor surface. Sample analyte is introduced so that binding interactions to the receptors on the cell surface can be studied. However, at times the analyte may interact nonspecifically with secondary targets such as non-cell sensor regions, the cell membrane wall, or alternative biomolecules on the cell membrane surface. This phenomenon is called nonspecific interaction. These nonspecific interactions are mainly due to a complex aggregate of molecular forces like charge interactions, hydrogen bonding and hydrophobic interactions. Unlike the affinity-based molecular interactions of interest, nonspecific interactions do not typically follow the law of mass action1 and can thus be readily identified with SPRM. This application note will address a variety of techniques commonly used to reduce the spurious effects of nonspecific interactions, as well as some techniques that are unique to SPRM.
I. Testing conditions
It is critical to know the isoelectric point, charge, composition (hydrophilic, hydrophobic) and size of the sample analyte in order to determine the appropriate testing conditions for reducing nonspecific interactions.
Buffer pH
The pH of the buffer used in binding interaction experiments has a significant effect on nonspecific interactions as it dictates the overall charge of the biomolecules in the experiment. For most experiments, pH should be close to the physiological pH (7-7.5). At this pH, the charge of the biomolecules can be estimated to determine if significant charge repulsion or attraction forces are present and whether these charging effects need to be mitigated.
Buffer salt concentration
Increasing the salt concentration in the running and sample buffers is a simple way to reduce nonspecific interactions. A high salt concentration increases the screening effect between charges. Salts such as NaCl prevent charges from the sample analyte from nonspecifically interacting with charges on the cell or sensor surfaces. For example, phosphate buffer saline is commonly used for cellular studies. The concentration can be increased up to 10x to avoid nonspecific interactions.
Buffer additives
Bovine serum albumin (BSA) protein is commonly added to the buffer in biosensor experiments to reduce nonspecific interactions. BSA is a globular protein with hydrophilic and hydrophobic subgroups which sacrificially pre-coat the sensor and cell hydrophobic regions thereby reducing nonspecific interaction events. Concentrations of BSA in the buffer are usually 0.05% to 0.3%.
Surfactants like Tween20 are also commonly added to the buffer of biosensor experiments. Tween20 disrupts hydrophobic interactions in the experimental environment thereby reducing nonspecific interaction events. It is important to note that the concentration of the surfactant should be adjusted according to the experimental conditions to avoid cell lysis. Tween20 concentrations up to 0.2% are routinely implemented.
II. Cell selection and growth conditions
Careful selection of cell types, growth conditions, and referencing methods can greatly help to reduce nonspecific interactions.
Adhesion layer
A variety of adherent layers may be applied to the biosensor surface to foster cell adhesion. Such adhesion layers act as a skeletal support for cell growth and typically consist of natural or synthetic polymers such as collagen, fibronectin or polylysine. Depending upon the type of sample being tested, some adhesion layers may perform better than otters at reducing nonspecific interactions. For example, collagen generally results in fewer nonspecific interactions than Poly D-Lysine (PDL) or Poly L-Lysine (PLL) for most types of antibody/cell interaction studies. However, for lectin/cell interaction studies collagen could produce more nonspecific interactions as it is a glycoprotein, unlike the synthetic polymers PDL and PLL2.
Growth media management
Fetal bovine serum (FBS) is a widely used growth supplement for cell culture of eukaryotic cells. FBS contains thousands of ingredients including proteins, electrolytes, lipids, carbohydrates, hormones, enzymes, and other undefined constituents, which are necessary in many culture conditions to support cell growth. FBS protects cells from large pH shifts, reduces toxicity, reduces the effect of proteases and helps to maintain a monolayer of cells during culture. Cell growth is fast and consistent in the presence of FBS. In spite of the variety of advantages of using FBS for cell culture, it can also be a significant reason for nonspecific interactions. Discussed below are a few effective ways to mitigate the nonspecific interactions caused by FBS.
A. Starving cells
Though most adherent cells require FBS in the media after seeding for growth, many cell lines can continue to grow even if the medium is devoid of FBS for 24 hours3. In such cases, serum free media may not be a significant reason for concern when the doubling time and viability remain satisfactory. This method of FBS starvation may greatly reduce nonspecific interaction by preventing the exposure of FBS to the biosensing area.
B. Suspension cells
A majority of the cells derived from vertebrates are anchorage-dependent (adherent cells) and must be cultured on a suitable surface to allow cell adhesion and spreading. However, many cell lines have been adapted for suspension-phase culture. Suspension cells are easier to culture as they grow in suspension and do not require enzymatic or mechanical forces to dissociate them from the culture flask. Once the grown suspension cells are rinsed free of growth media, they can be directly settled onto the biosensor surface for immediate testing. This approach has a major advantage for reducing nonspecific interactions in that the exposure of FBS to the biosensor surface is prevented.
Additionally, BI’s SPRM technology is unique in its ability to facilitate suspension cell based interaction studies. Its simultaneous brightfield and SPR imaging capability of the biosensing area enables accurate monitoring of the suspension cell settling process. SPRM is not only able to visually observe in real-time the number of cells on the biosensor surface, but also simultaneously determine the strength of each cell’s capture to the biosensor surface based upon the localized intensity change of the surface plasmon resonance signal. In this way, one can more accurately determine the confluency of stably-bound cells before removing the suspension of cells. Once the desired cell confluency is reached (typically 50 to 80%), the cell suspension solution can be removed to terminate the cell capture process and to begin the sample binding interaction study. Human Burkitt’s Lymphoma B suspension cell (Ramos cells) interactions between lectins like Wheat germ agglutinin (WGA) and glycoproteins present on the membrane have been studied in this way, exhibiting very low nonspecific interactions4.
C. Surface patterning
Surface patterning techniques may be implemented by image-based biosensor technologies such as SPRM for the reduction of nonspecific interactions. In this approach, specific areas on the biosensor surface may be designated for cell growth or for referencing. For example, FBS exposure can be prevented in some areas by using a temporary surface blocker ( e.g. a stamp). The stamp is attached to a portion of the biosensor surface protecting it from FBS exposure during cell growth as seen in Figure1. After cell growth, the stamp is removed, resulting in a FBS-free area for referencing.
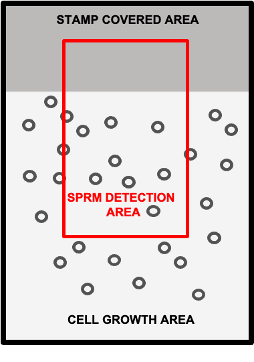
Figure 1: Schematic drawing of a biosensor chip containing a FBS-free region that was previously protected by a stamp, and a neighboring region in which cells were permitted to grow freely. The red square indicates an example area for detection that includes both cell regions and FBS-free regions for referencing.
III. Data processing and analysis
The data processing and analysis capability of BI’s SPRm200 excels in its ability to reduce nonspecific interactions to produce relevant and high quality binding interaction results.
Reference subtraction
The subtraction of a reference area helps to improve data quality by removing common noise and secondary effects such as nonspecific interactions. The careful design and selection of a reference is critical to the effectiveness of reference subtraction. Since BI’s SPRm200 uses a high resolution image-based biosensor technology, it has the luxury of utilizing a variety of types of regions on the sensor as references including pre-patterned areas 5, bare sensor areas, the tiny areas in between cells, and even other cells as references.
BI’s ImageSPR™ software (included with SPRm200) can automatically identify cell and non-cell areas on the biosensor surface, and then evaluate multiple types of references to reduce nonspecific interactions for kinetic interaction analysis.
Tightening fits
BI’s ImageSPR™ software provides statistical analysis of the measured binding affinity and kinetic constants. By tightening the fitting-error tolerance and narrowing the distribution of interaction results, more of the poorly or oddly responding activity regions are eliminated. Such regions are mostly attributed to nonspecific interaction behavior. In this way, the interaction analysis becomes more meaningful, greatly improving the quality of the results as seen in Figure 2.
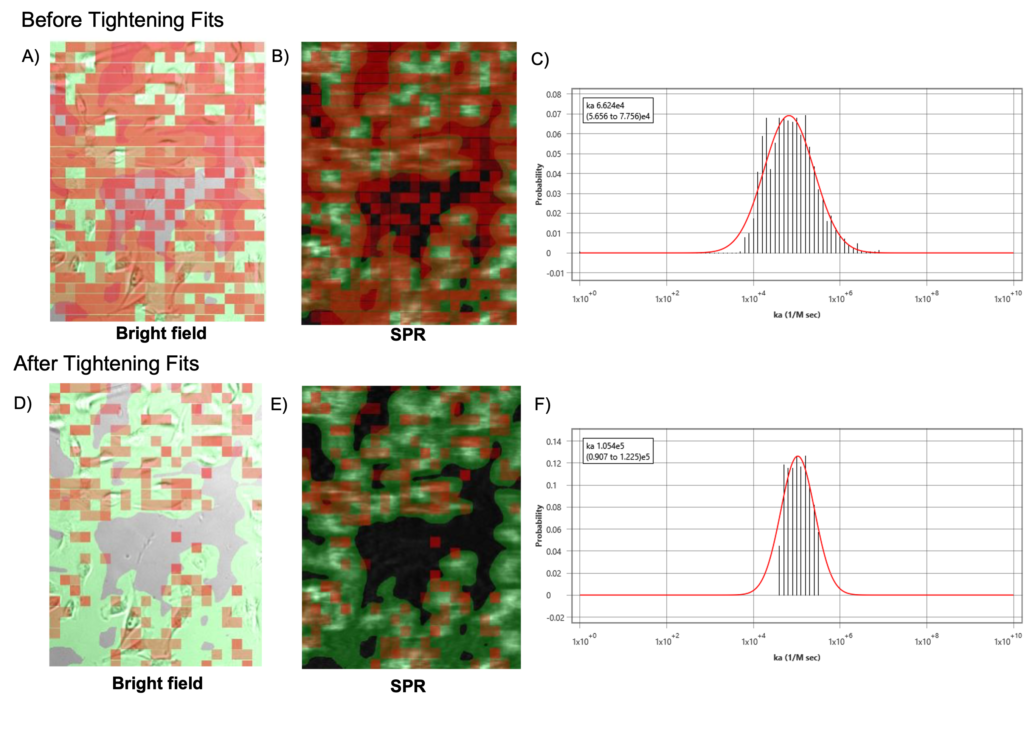
Nonspecific interactions are problematic for all biosensor technologies, as they interfere with the affinity binding response. A variety of methods to reduce nonspecific interactions were presented. Several methods such as precision identification of cell and reference areas, utilizing multiple types of references for subtraction, and statistics-based interaction analysis exemplify how BI’s SPRM technology excels in its ability to reduce nonspecific binding, and thereby produce relevant high-quality binding interaction results.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published June 5, 2023
DOWNLOAD PDF
Download a PDF of Application Note 146: ENHANCED REDUCTION OF NONSPECIFIC INTERACTIONS
- Frutiger, Andreas, et al. "Nonspecific binding—fundamental concepts and consequences for biosensing applications." Chemical Reviews 121.13 (2021): 8095-8160
- Varki, A., et al. "Antibodies and lectins in glycan analysis." (2009).
- Klasić, Marija, et al. "DNA hypomethylation upregulates expression of the MGAT3 gene in HepG2 cells and leads to changes in N-glycosylation of secreted glycoproteins." Scientific Reports 6.1 (2016): 24363.
- SPR Microscopy for Live Suspension Cells, App note #131 (https://biosensingusa.com/application-notes/).
- Han, Chaowei, et al. "Microfluidically Partitioned Dual Channels for Accurate Background Subtraction in Cellular Binding Studies by Surface Plasmon Resonance Microscopy." Analytical Chemistry 94.49 (2022): 17303-17311.
