Cell-based immunotherapy has gained great attention from researchers and pharmaceutical companies, particularly due to its promise to treat various cancers. Understanding the interaction between drug candidates and targets on the cell membrane is crucial to drug development. One area of particular importance is the interaction between drugs and immune cells. However, this is a complex biological process, which not only depends on the drug molecules and receptors engaged, but also on the possible clustering and dimerization of the receptors induced by the molecular binding. Techniques used for studying the interaction between drug molecules and their targets often require the target proteins be extracted from the cell or the cells being fixed. Unfortunately, the integrity of a live cell could be compromised by protein extraction and cell fixation. Therefore, it is advantageous to directly characterize the molecular interaction on living cells.
SPR microscopy (SPRM) has been demonstrated as a viable technique capable of measuring binding affinity and kinetics at cell membranes. We demonstrate in this application note that SPRM is capable of performing kinetic studies of living cells. Compared to instruments designed for studies of live cells, such as FACS and LigandTracer, SPRM is a label-free technique for measuring binding kinetics and affinity in real time. The system integrates SPR and optical microscopy for simultaneous cell imaging and SPR binding measurements.
In here, we studied the binding of Rituximab, a mouse/human chimeric monoclonal antibody, to the CD20 receptors, which are endogenously expressed by B cells. This antibody was approved by the US FDA in 1997 to treat B-cell non-Hodgkin lymphomas. It acts by depleting normal as well as pathogenic B cells while sparing plasma cells and hematopoietic stem cells, which do not express the CD20 surface antigen.
Rituximab was purchased from InvivoGen. Ramos cells, purchased from ATCC, were cultured in a growth medium before being transferred to about 1 mL of imaging buffer at a density of about 1 x 106 cells/mL. This cell imaging buffer containing 0.1% BSA was also used as the running buffer. Au sensor chips coated with PLL were used as the substrate for capturing and anchoring the B cells. About 50 uL Ramos cell solution was pipetted onto the chip. Capture of the Ramos cells was simultaneously monitored by the bright-field camera and SPRM. Additional aliquots of the Ramos cell solution can be added onto the chip if a greater confluence is desired. Each chip was washed with running buffer to remove excessive cells before mounting onto the SPRM. Eight serial dilutions were made from a 76.7 nM stock in running buffer (x3 dilution) and injected into the system using the kinetic titration method.
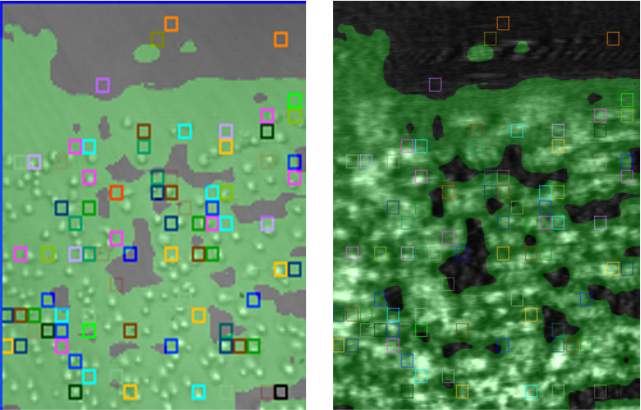
Captured Ramos cells were stable on the PLL-coated chip surface during the experiment. As seen in the bright field image in Fig.2, they maintained their spherical shape with no obvious degradation. The statistical analysis of binding affinity and kinetic constants reveals the heterogeneity between cells and the averaged kinetic constants. The colored squares indicate the valid ROIs (regions of interest) used for the statistical calculation of the kinetic parameters, which overlap well with the actual positions of the attached cells.
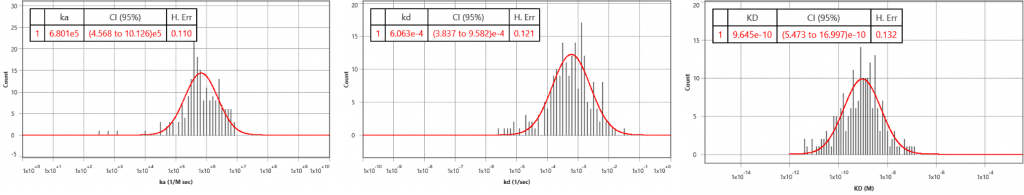
The average equilibrium and kinetic constants are KD = 965 pM, ka = 6.8 x 105 M-1 s-1 and kd = 6.0 x 10-4 s-1. These values are in close agreement with reported values (~1 nM) from other measurements and that cited by FDA (5.2-11 nM). The statistical distribution of these values partially reflects the dynamic complexity of the living system that affect the binding process.
In summary, SPRM is capable of measuring binding affinity and kinetic constants of membrane targets at living cells, which are difficult to obtain using traditionally label-based and label-free methods. Measuring biophysical properties of a target in the native membrane environment in real time without labels provides more accurate information about the drug-receptor interactions, opening an opportunity to speed up drug development, particularly for molecularly targeted therapy.
We thank Nanxi Yu from ASU for assisting with the samples and Jon Brooks from Pfizer for the valuable discussions.
DOWNLOAD PDF
Download a PDF of Application Note 141: SPRM Measurements of Binding Kinetics between Rituximab and CD20 on Live B Cells
