Surface Plasmon Resonance (SPR) is a powerful technique for measuring the binding kinetics of biomolecular interactions in a real-time and label-free manner [1]. In traditional SPR assays, the target molecule is extracted and purified from the cell and immobilized onto the sensor surface for measurement of the interaction kinetics between the target and the drug candidate. The collection and purification of the biomolecules from the cells are not only time consuming, but also changes the native environment of the target. The latter is particularly problematic for membrane proteins, as they require a lipid bilayer environment to maintain their integrity and stability [2]. Surface Plasmon Resonance Microscopy is a new technique that enables measurement of binding kinetics of membrane proteins in vitro and therefore in their true native membrane environment [3].
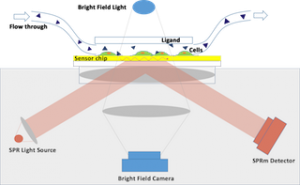
The SPRm 200 from Biosensing Instrument Inc. is a system that seamlessly combines both optical bright-field and SPR microscopies thus enabling the unique in vitro binding kinetic measurement of individual cells in their native environment. The architecture of the system consists of using the SPR phenomenon to measure binding kinetics while also simultaneously permitting use of bright-field microscopy to study topographical and phenotypical changes in the sample (Fig. 1). Typically, the cells are cultured and fixed on a poly-L-lysine coated SPR sensor chip. The integrated optical microscope allows for visualization of the sample with x10 magnification and 1 um resolution. The SPRm 200 exceeds the performance of homemade systems by demonstrating a much larger field of view, simultaneous bright-field and SPR microscopies, and greater stability.
In this study, two types of cells, SH-EP1 human epithelial cells and Barrett’s esophagus-derived CP-D (CP- 18821) cells were used to measure the binding kinetics of glycoproteins. Glycoproteins are glycosylated membrane proteins that play an integral role in the cell-cell and cell-matrix recognition events [4]. The structure and type of sugar residue is critical to the protein’s function and is an area of significant research interest. Wheat-germ agglutinin (WGA, 36k Da) is a lectin protein that can recognize sugar residues on glycoproteins. When WGA is exposed to the cells an increase in SPR signal is observed indicating that the WGA lectin proteins are binding to the sugar residues of the cell membrane glycoproteins [5].
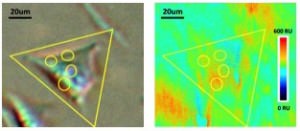
Sixteen SH-EP1 cells were selected for this study (Fig. 2). WGA in five different concentrations (12.5, 25, 50, 100, 200 ug ml-1) at a flow rate of 300 ul min-1 and a regeneration solution of 50 mM N- acetylglucosamine were used. Based on the 1:1 Langmuir model, the average ka (association), kd (dissociation) and KD (affinity) of these 16 cells were found to be 4.3 x 103 M-1 s-1, 1.1 x 10-3 s-1 and 257 nM respectively. These values are in good agreement with published literature.v Additionally, this technique allows researchers to study and characterize the heterogeneity of cellular populations since the binding responses of each cell may be individually observed.
SPRm 200 has the resolution to observe sub-cellular binding activities (Fig. 3). In this experiment, a single CP-D cell was used to study the binding with WGA. Different sub-regions of the cell were selected for study. The kinetic values of the different sub-regions on the cell were similar. However, the SPR response magnitudes varied depending upon the local surface density of the membrane receptors. The average KD for the cell was 24 nM, well in accordance with published materials [6]. The observed difference in the WGA binding interaction of glycoproteins on CP-D cells and SH-EP1 cells is attributed to differences in their sugar structures, and further illustrates the importance of this technique in studying the effect of post-translational modifications on membrane proteins in their native cellular environment.
In conclusion, SRPm 200 is a novel instrument that seamlessly integrates bright-field and SPR microscopies into one system. This instrument enables the kinetic binding study of multiple individual cells as well as the localized responses on a single cell.
Author: Nguyen Ly | Biosensing Instrument | Published May 4, 2020
DOWNLOAD PDF
Download a PDF of Application Note 122: Surface Plasmon Resonance Microscopy for Multiple and Single Cell Membrane Binding Kinetics Studies
- Schasfoort, Richard B M and Tudos, Anna (eds), RSC Publishing, 2008, 1-14
- Raschle, T., Hiller, S., Etzkorn, M. and Gerhard Wagner, G., Non-micellar systems for solution NMR spectroscopy of membrane proteins, Curr Opin Struct Biol. 20, 471–479 (2010)
- Patching, S., Surface Plasmon Resonance Spectroscopy for Characterization of Membrane Protein-ligand interactions and its potential for Drug Discovery, Biochimica et Biophysica Acta 1838, 43-55 (2014)
- Dell, A. and Morris, HR, Glycoprotein structure determination by mass spectrometry, Science 291, 2351-2356 (2001)
- Wang, W., Yang Y., Wang, S., Nagaraj, V., Liu, O., Wu, J.,Tao, N.J., Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells, Nature Chemistry 4, 846–853 (2012)
- Guan, Y., Shan, X., Zhang, F., Wang, S., Chen, H., Tao, N.J., Kinetics of small molecule interactions with membrane proteins in single cells measured with mechanical amplification, Science Advances, e1500633 (2015)