Surface Plasmon Resonance (SPR) is a widely used label-free detection technique for studying binding behavior of biomolecules. Since its commercialization in 1990’s [1], SPR has made vast advances in terms of both development of the technology and its applications. It has become a central tool in biomedical research, biosensor development and drug discovery. Over the last ten years, SPR has also been coupled with other techniques such as electrochemistry (EC-SPR) [2], HPLC, and mass spectrometry (SPR-MS) [3], and has been used for trace detection in the gas phase (SPR-gas detection). One of the most significant developments in recent years is SPR microscopy (SPRM), a technique that combines high-resolution optical microscopy with SPR technology. SPRM provides the benefits of both techniques and offers direct spatial visualization of samples and real-time measurements of binding affinity and kinetics. It has been challenging to study cell membrane protein binding with drug candidates or other ligands for conventional (channel based) SPR because the extraction of proteins from cells and immobilization of purified proteins onto the SPR sensor are not only laborious but also change the native environment of the proteins. SPRM has significantly improved such measurements by affixing cells directly on the SPR sensor, thereby keeping proteins in their native states [4]. Furthermore, distribution and local binding activities of membrane proteins in each cell can be determined and mapped.
In this technical note, SPRM technique is reviewed and compared to conventional SPR.
Conventional (channel based) SPR
The most common SPR mode for molecular binding studies is based on fluidic channels (Figure 1). A microfluidic module with multiple channels is mounted on top of a sensor and samples are delivered by a pumping system into each channel. Ligand molecules are immobilized onto sensing area of each channel, and the analyte molecules are subsequently delivered into each channel to bind with a specific ligand. Data are averaged over the sensing area for each channel (purple-colored area) to yield SPR sensorgrams (shown on the right of Figure 1). Each channel generates a sensorgram (or several sensorgrams after surface regenerations) during the measurement and binding affinity and kinetic parameters are derived from these sensorgrams.
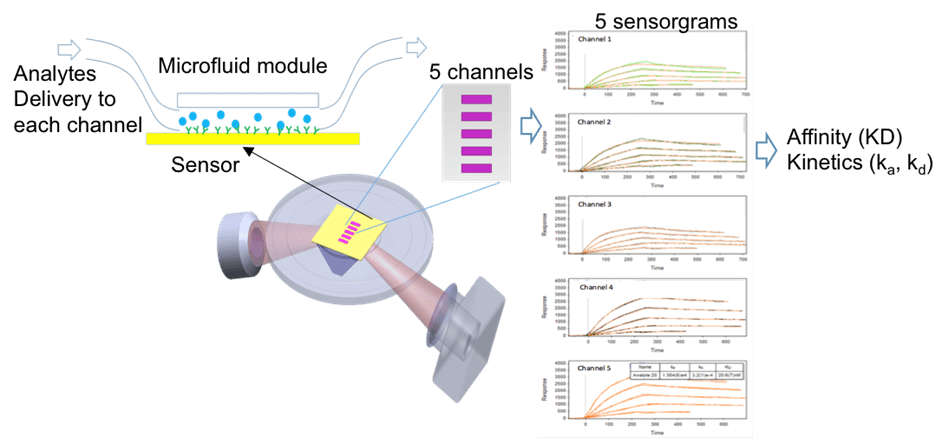
FIG. 1 Conventional (channel-based) SPR mode
SPRM (high-resolution microscopy)
SPRM is a high-resolution imaging-based SPR mode. It provides both bright field optical images and SPR data (affinity and kinetic parameters) of the sensing area (Figure 2). SPRM (such as SPRm200) produces one SPR sensorgram at each pixel of the image, offering spatial mapping of the binding activities over any given sensing area and much more information than the conventional SPR mode. SPRM can be used not only for conventional SPR measurements of binding analysis, but also for heterogeneity studies of cellular response to drug candidates. Because of its high spatial resolution (~ 1 µm), it can monitor and measure virus and bacteria binding activities, which cannot be directly measured with the conventional SPR mode. SPRM is especially powerful in measuring drug interactions with membrane proteins in their native state as the protein molecules are within the membranes of live cells.
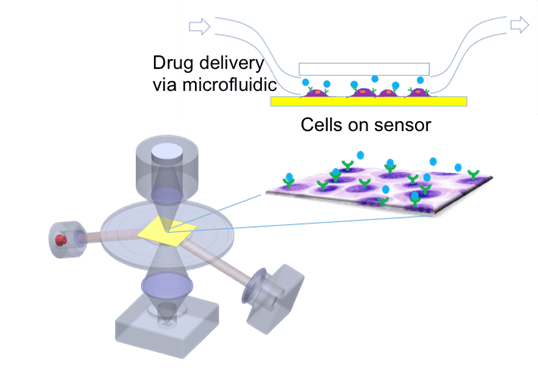
FIG. 2. High-resolution imaging-based SPR mode
Cells are incubated or immobilized on the sensor surface, and the analyte or drug molecules are delivered to the sensing area. Figure 3 shows an example of an interaction between drug molecules and cells incubated on a sensor. The real-time SPR image (3B, blue image) indicates the interactions or binding response (pink area) of cell receptors to the drug molecules. By selecting the areas of interest based on a bright field image (3A, green image), sensorgrams can be generated (3C) and binding information can be derived. When many different regions of cells are chosen, the heterogeneity of the cell and cell membrane can be determined with statistically meaningful data.
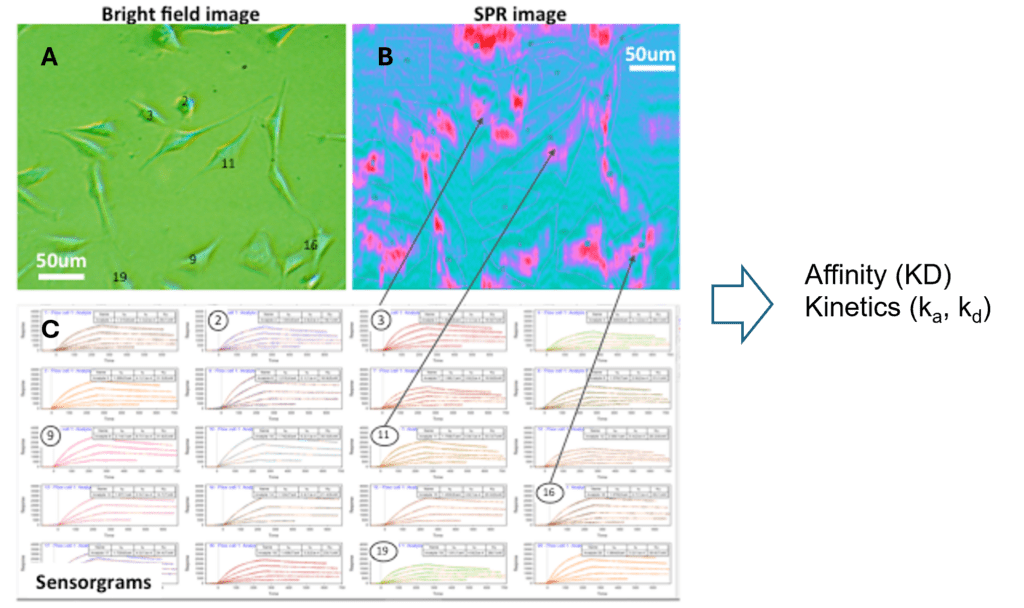
FIG. 3 SPRM provides simultaneous bright field image (A) and SPR image (B). Binding parameters are derived by the sensorgrams (C).
In summary, SPRM provides a unique capability to visualize the binding activities of cell membranes. Compared to the conventional channel-based SPR mode, SPRM has the added advantage of spatial mapping of the binding affinity and kinetics of cell membrane proteins in their native state, thus providing more biologically relevant information. Due to its high spatial resolution, SPRM can be used to monitor other surface processes such as binding of nanoparticles tagged with biomolecules or delivery of drugs encapsulated by nanoparticles. (Please visit our webinar series page for various applications)
The table below summarizes the key points of SPRM in comparison with SPR.
| SPR | SPRM (SPRm200) | |
| Detection technique | SPR | SPR microscopy and optical imaging |
| Sensor area | Channel area averaged | Per pixel |
| Sample delivery | Channel based microfluidic delivery | Microfluidic delivery |
| Sample type | Drug molecules Protein Lipid DNA | Drug molecules Protein Lipid DNA Cell Virus Bacteria Nano-particles |
| Data type | Sensorgram curves per channel | Sensorgram curves per pixel Bright field optical image with simultaneous SPR images |
| Measured quantities | Affinity Kinetic constants | Affinity Kinetic constants Heterogeneity study of cellular response Nanoparticle activities Bright field image |
Check out our SPRm200 video here for more general information.
Authors: Nguyen Ly | Biosensing Instrument | Published May 22, 2017
