Antibody drug development is emerging as the cornerstone of modern therapeutics, particularly in the realm of immunotherapy. Antibodies have demonstrated great efficacy in treating B-cell cancers by taking advantage of highly specific interactions with cell surface receptors. Proper comprehension of the binding kinetics of the antibodies to target cells is central to optimizing the prediction of in vivo performance and overall therapeutic efficacy.1 While traditional analyses like ELISA have been focused on immobilized targets or recombinant proteins, quantitating antibody interactions in live suspension cells provides physiological insight. Live suspension B cells preserve native antigen presentation, membrane fluidity, and cell dynamics critical to accurately measure drug targeting. Studying these interactions in real time offers a direct window into how various antibody base therapeutics perform in a biological context.2
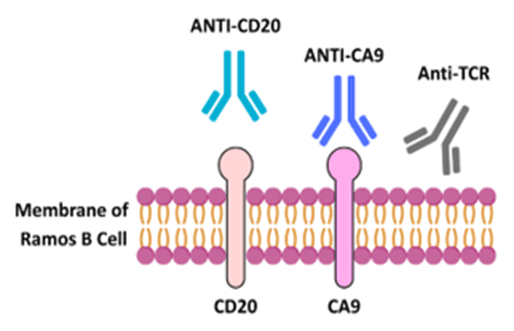
Figure 1: Schematic of antibody binding to the receptors in the membrane of Ramos B cells.
Surface Plasmon Resonance Microscopy (SPRM) has emerged as a highly capable technology for label-free real-time examination of molecular interactions on live cells. By presenting kinetic profiling in native cell conditions at high resolution, SPRM bridges the gap between in vitro biochemical assays and functional in vivo results.3 In this study, we used SPRM to monitor the kinetic binding of three different monoclonal antibodies: anti-CD20, anti-CA9, and anti-TCR onto live suspension Ramos B cells, a human Burkitt lymphoma cell line. This approach allows direct measurement of antibody binding to native membrane architecture, receptor accessibility variation, and intrinsic cell-to-cell heterogeneity.
Anti-CD20 binds to the hallmark B cell surface antigen CD20, strongly and stably expressed on mature Ramos B cells. Its selective expression on B cells and absence of expression on plasma cells and other immune lineages positions CD20 as an ideal therapeutic target for monoclonal antibodies such as Rituximab.4 Anti-CA9 is directed against Carbonic Anhydrase IX, a hypoxia-regulated membrane protein that is also commonly overexpressed in solid tumors but presents at moderate levels in B cells compared to the robust CD-20 receptor.5 Anti-TCR, which binds to the T cell receptor, was included as a negative control to assure assay specificity since TCR is not present in B cells.6 Binding comparison on the same live cell population of all three antibodies permits direct, physiologically relevant comparison of binding performance and offers a window into heterogeneous antigen expression at the cellular level that is incredibly valuable for the evaluation of therapeutic targeting in complex tissues.
Although both anti-CD20 and anti-CA9 are in vitro high-affinity antibodies, their binding property on living Ramos cells was vastly different. Anti-CD20 showed a high binding activity consistent with the high surface expression and accessibility on Ramos B cells with an affinity of 5.1 nM (Figure 2a). On the other hand, anti-CA9 had comparatively weaker binding of high affinity 1.2 nM, possibly due to lower total receptor density (Figure 2b). Moreover, some cells may express no or minimal CA9, limiting the total signal and affecting the apparent kinetic profile. Anti-TCR exhibited no measurable binding, which ensured the absence of its target on Ramos cells and reinforced the SPRM’s ability to distinguish true biological interactions from nonspecific background (Figure 2c).
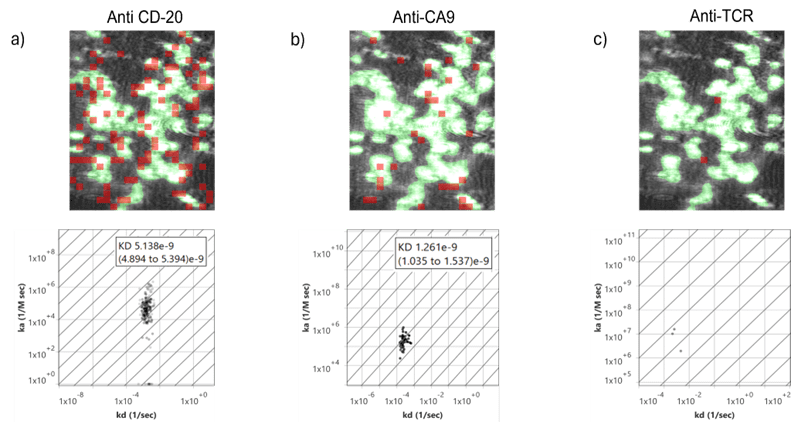
Figure 2: SPR images displaying the cell regions highlighted in green with red highlights indicating areas of detected a) anti-CD20, b) anti- CA9 and c) anti-TCR binding interactions with corresponding affinity isotherm plot extracted from hundreds of responsive ROIs showing total kinetic interactions for three antibody interactions on live suspension Ramos B cells.
These differences point to the importance of studying antibody binding in a live cell, where kinetics of local microenvironment dynamics can affect interaction rates in ways not captured by traditional end point assays. These findings illustrate the key importance of membrane presentation and expression heterogeneity in determining functional antibody behavior. Use of SPRM to determine antibody binding to CA9, CD20, and TCR in live Ramos B cells demonstrates its unique ability to capture biologically meaningful kinetics. It not only confirms specificity of binding but also reveals differences in target interaction that have the potential to guide the outcome in therapeutics. Lastly, SPRM drives the development of future-generation antibody therapeutics by giving insights that are in harmony with clinical reality that do not come from other conventional means.
Author: Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published July 1st, 2025
DOWNLOAD PDF
Download a PDF of Application Note 167: Label-Free Characterization of Antibody-Receptor Interactions on Live Suspension Cells using SPRM
- Chan, Andrew C., and Paul J. Carter. "Therapeutic antibodies for autoimmunity and inflammation." Nature reviews immunology 10, no. 5 (2010): 301-316.
- Jain, Maneesh, Neel Kamal, and Surinder K. Batra. "Engineering antibodies for clinical applications." Trends in biotechnology 25, no. 7 (2007): 307-316.
- Wang, Wei, Yunze Yang, Shaopeng Wang, Vinay J. Nagaraj, Qiang Liu, Jie Wu, and Nongjian Tao. "Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells." Nature chemistry 4, no. 10 (2012): 846-853.
- Wojciechowski, Wojciech, Huifen Li, Shannon Marshall, Chiara Dell’Agnola, and Igor Espinoza-Delgado. "Enhanced expression of CD20 in human tumor B cells is controlled through ERK-dependent mechanisms." The Journal of Immunology 174, no. 12 (2005): 7859-7868.
- Sedlakova, Olga, Eliska Svastova, Martina Takacova, Juraj Kopacek, Jaromir Pastorek, and Silvia Pastorekova. "Carbonic anhydrase IX, a hypoxia-induced catalytic component of the pH regulating machinery in tumors." Frontiers in physiology 4 (2014): 400.
- Rappazzo, C. Garrett, Monica L. Fernández-Quintero, Andreas Mayer, Nicholas C. Wu, Victor Greiff, and Jenna J. Guthmiller. "Defining and studying B cell receptor and TCR interactions." The Journal of Immunology 211, no. 3 (2023): 311-322.
