SPR (SPR) has been demonstrated as a powerful technique for rapid, sensitive, and label-free genetic analysis [1-5]. When the sensor surface is coated with a single sensing (probe) DNA, SPR can be used for both affinity binding studies (i.e., kinetic measurements) and concentration detection of a target DNA. However, the concentration detection levels for SPR and SPR imaging are typically at low nanomolar (nM) [3, 4], which cannot rival fluorescence measurements. The remarkable sensitivity of SPR based on a position-sensitive device, on the other hand, enables researchers to measure oligonucleotide (ODN) and polynucleotide targets at sub-picomolar (sub-pM) [1]. Recently, low femtomolar level detections have even been achieved by utilizing ODN-capped gold nanoparticles [2] and enzyme-catalyzed reactions [4, 5]. This application note describes the use of a Biosensing Instrument (BI) SPR instrument, in conjunction with an enzyme-catalyzed precipitation reaction, to achieve highly sensitive and sequence-specific DNA assays.
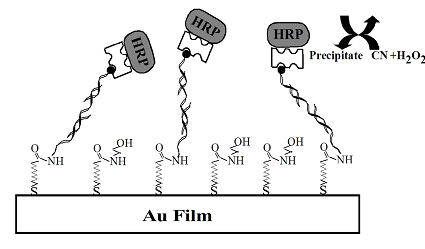
FIG. 1 Schematic scheme for the sandwich DNA assay amplified by precipitate formed from enzymatic reactions. For clarity, the molecules are not drawn to scale.
To measure the DNA targets at ultratrace levels, aminated ODN capture probes were affixed onto a carboxylated self-assembled monolayer preformed onto gold SPR sensor chips (available from Biosensing Instrument) via the well-known EDC/NHS (1-(3-dimethylaminopropyl)-3-ethyl-carbodiimidehydrochloride/N-hydroxysuccinimide) cross-linking reaction (FIG. 1). Upon hybridization with DNA targets of different concentrations or in various samples, the sensor surface was exposed to a biotinylated detection probe. This was followed by the attachment of the horseradish peroxydase-tagged streptavidin. The resultant sensor chip was mounted onto the prism of a BI-SPR 1000 instrument. A PBS (10.0 mM phosphate, pH = 7.0, and 0.1 M NaCl) solution was delivered by a syringe pump to the dual-channel PEEK (polyetheretherketone) flow cell equipped with a PDMS (polydimethylsiloxane) gasket. In this setup, one channel can be employed for sample analysis while the other serves as the reference. Alternatively, both channels can be used to perform replicate analyses or measurements of different samples. For the amplified SPR gene detection shown in FIG. 1, a six-port, through-the-handle injection valve was used to inject hydrogen peroxide (H2O2) and 4-chloro-1-naphthol (CN) loaded in a 50-µL sample loop into the flow cell. As depicted in FIG. 1, the enzyme, horseradish peroxydase, anchored onto the sensor surface due to sequence-specific DNA hybridization, catalyzes the reduction of CN in the presence of H2O2.[5] A precipitate produced from this reduction deposits onto the sensor surface, resulting in a considerable change in the SPR signal.
FIG. 2 is an overlay of five representative SPR sensorgrams showing the sequence specificity of the analysis. For the 33-mer target, the SPR signal corresponding to the hybridization reaction between the capture probe and its complementary target (plateau of curve d) decreased by more than 50% when the target has only one base mismatching the capture probe (curve c). This suggests that, when surface modification and DNA hybridization conditions are optimized, the SPR technique can provide a viable means for single-base mismatch assay (i.e., SPR is amenable to single-nucleotide polymorphism study). The presence of four mismatching bases on the target further decreased the signal (curve b), and very little SPR signal was observed when a non-complementary target was injected (curve a). Another control experiment (curve labeled with “control”) involved injecting the mixture of H2O2 and CN into the flow cell housing a sensor chip covered with the DNA duplexes that do not contain the biotin tags. As a result, HRP could not be attached to the sensor surface and no precipitate would deposit. This is evidenced by the steady baseline in the control curve.
FIG. 2 SPR sensorgrams recorded upon injecting 1 mM CN and 0.3 mM H2O2 onto the surfaces that have been treated with four ODN targets with different sequences: a non-complementary sequence to the capture probe (a), four bases mismatching to the capture probe (b), one base mismatching to the capture probe (c), and a complementary sequence to the capture probe (d). The control sensorgram was obtained at a sensor chip covered with the capture probe/target duplex that had been exposed to the HRP-tagged streptavidin. The concentration of the target used was 50 fM. The arrow indicates when the injections were made.
The concentration detection levels using the BI-SPR instrument in tandem with the signal amplification are quite noteworthy. The plot of SPR signal vs. ODN target concentration (not shown here) exhibits two regions. The lower curve (from 10 fM to 5 pM) has a slope that is much steeper than that of the upper section (from 5 pM to 100 pM). The variations in the slopes (sensitivities) between the lower and upper regions can be interpreted on the basis of the surface coverage of the duplexes formed at different target concentrations. Thus, ODN concentrations as low as a few femtomolar can be measured. Such remarkably low detection levels can be attributed to the combination of this highly sensitive SPR instrument and the signal amplification scheme.
Author: Nguyen Ly | Biosensing Instrument | Published June 11, 2014
DOWNLOAD PDF
Download a PDF of Application Note 106: Flow Injection-SPR: Sensitive and Sequence-Specific DNA Assays
- Song, F., Zhou, F., Wang, J., Tao, N., Lin, J., Vellanoweth, R. L., Y.;, M., and Laidman, J. W. Nucleic Acids Res. 2002, 30, e72
- X. Yao, X. Li, F. Toledo, C. Zurita-Lopez, M.Gutova, J. Momand, F. Zhou, Anal. Biochem., 2006, 354, 220-228
- He, L., Musick, M. D., Nicewarner, S. R., Salinas, F. G., Benkovic, S. J., Natan, M. J., and Keating, C. D., J. Am. Chem. Soc., 2000, 122, 9071-9077
- Lee, H. J., Li, Y., Wark, A. W., and Corn, R. M., Anal. Chem. 2005, 77, 5096-5100
- Yin, L., Zhang, Y., Zeng, D., Wang J., Zhou F. Frontier of Bioscience 2007, in press
- L. Alfonta, A. Singh I. Willner, Anal. Chem. 2001, 73, 91-102
