Mucin-4 (Muc-4) is a heavily glycosylated membrane glycoprotein which is associated with pancreatic cancer and metastasis.1 This glycoprotein primarily has O-glycans which contributes to its bulky structure in the extracellular region and some N-glycosylation sites that are part of the transmembrane region.2 Also, it is a novel tumor antigen that significantly contributes to pancreatic cancer development, which is absent in the normal pancreas, making it a highly attractive candidate for immunotherapy and vaccine development. Additionally, the aberrant glycosylation within and around Muc-4, as shown in Figure 1, has shown to contribute to tumor growth.3&4 Therefore, understanding the influence of aberrant glycosylation on the binding capability of Muc-4 in the native microenvironment is crucial to test the effectiveness of new drugs.
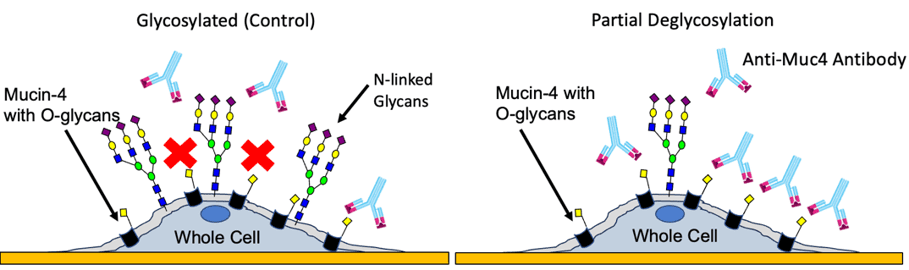
Figure 1: Schematic of anti Muc-4 antibody interactions with Muc-4 in glycosylated and partially deglycosylated pancreatic cancer cells.
In this study, Surface Plasmon Resonance (SPR) Microscopy was implemented to study complex influences of the native N-glycan cellular environment on binding interactions to the Muc-4 receptor as this is currently the only commercially available label-free technique with high enough sensitivity and resolution to measure binding kinetics and heterogeneity on single cells.5 Such unique capability enables for a more accurate understanding of the “true” binding interactions on human cancer cells without disrupting the native environment of the target Muc-4 receptor.5
To understand the influence of glycosylation on Muc-4 in the cellular environment, the binding interactions of monoclonal antibody anti-Muc-4, which targets the extracellular region of Muc-4, was studied using our SPRm 200 instrument. The kinetic results reveal two distinct anti-Muc-4 binding interaction modes in glycosylated BxPC3 cells (Fig 2A and 2B, Table 1), with mode (a) having a 4x faster on-rate and higher affinity than the other mode (b).
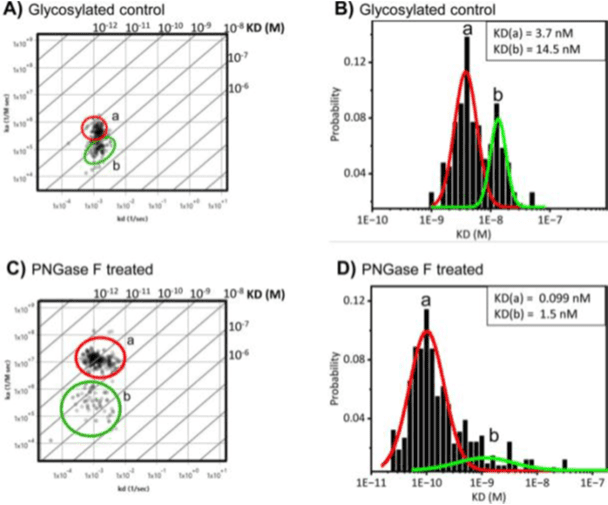
Figure 2: Binding affinity of Anti-Muc-4 monoclonal antibody increases on deglycosylated BxPC3 pancreatic cancer cells. A) Affinity isotherm plot extracted from hundreds of responsive ROIs for anti-Muc-4 on glycosylated BxPC3 cells which produces two distinct binding interactions, labeled as modes a and b. B) Histograms describing kinetic interactions and distributions for anti-Muc-4 on glycosylated BxPC3 cells. C) Affinity isotherm plot extracted from hundreds of responsive ROIs for anti-Muc-4 on deglycosylated BxPC3 cells which produces higher affinity and faster association for the two binding interaction modes labeled as modes a and b. D) Histograms describing kinetic interactions and distributions for anti-Muc-4 on N-linked deglycosylated BxPC3 cells. a and b represent binding interaction modes. Histogram data were graphed and fitted using Origin 2023b. Affinity isotherm plots were acquired using BI’s Image SPR software.
| Cells | KD (nM) | 95% Cl (nM) | ka (M-1s-1) | ka (s-1) |
| Glycosylated BXPC3 cells | a) 3.7 b) 14.5 | a) 3.2 to 4.1 b) 13 to 15 | a) 4.3X105 b) 1.2X105 | a) 1.5X10-3 b) 1.4X10-3 |
| Deglycosylated BXPC3 cells | a) 0.099 b) 1.5 | a) 0.082 to 0.10 b) 1.3 to 2.4 | a) 1.1X107 b) 7.0X105 | a) 1.0X10-3 b) 1.1X10-3 |
Table 1: Kinetic parameters of anti Muc-4 binding to Muc-4 on BxPC3 cells. All data are representative from 5 different experiments. A 1:1 binding model was applied to all Muc-4 data analysis.
The extracellular domain of Muc-4 is exclusively O-linked glycosylated, making the presence of any N-linked glycoforms in this region the likely result of encroachment from neighboring glycoproteins.
Upon partial removal of N-linked glycans with PNGase F enzyme in PC cells, two distinct binding interaction modes are still observed. However, mode (a) shifted to ~25x faster on-rate and 37x higher affinity relative to the glycosylated control mode (a). Mode (b) displayed a much broader distribution with a low probability, and a ~9x higher affinity relative to the glycosylated control mode (b) (Figure 2, Table 1). The second binding mode b is attributed to be a hindered mode. This is supported by the observation that after partial N-linked glycan deglycosylation, the binding occurrences of mode b are significantly diminished, whereas those of mode a are significantly more abundant (Figure 2C and 2D).
SPR Microscopy allows for a real-time, accurate understanding of binding interactions of various potential therapeutic drug targets in the native heterogenous cellular environments. These SPR Microscopy results reveal the complex influence of extraneous N-linked glycans on the binding heterogeneity of Muc-4 of pancreatic cancer cells, which renders important implications to improve Muc-4 detection and its utilization as a potential biomarker of pancreatic cancer.
Author: Jesús Aguilar Díaz de león, Nguyen Ly and Miyuki Thirumurthy | Biosensing Instrument | Published September 24th, 2024
DOWNLOAD PDF
Download a PDF of Application Note 160: Influence of Aberrant Glycosylation on the Binding Capability of Muc-4 in Pancreatic Cancer Cells
- Chaturvedi P, Singh AP, Batra SK (2008) Structure, evolution, and biology of the Muc-4 mucin. The FASEB Journal 22:. pmid:18024835
- Dreyer CA, Vorst K Vander, Free S, Rowson-Hodel A, Carraway KL (2022) The role of membrane mucin Muc-4 in breast cancer metastasis. Endocr Relat Cancer 29
- Zhang Y, Sun L, Lei C, Li W, Han J, Zhang J, et al. (2022) A Sweet Warning: Mucin-Type O-Glycans in Cancer. Cells 11 https://doi.org/10.3390/cells11223666 PMID: 36429094
- Ho WL, Hsu WM, Huang MC, Kadomatsu K, Nakagawara A (2016) Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J Hematol Oncol 9 https://doi.org/10.1186/ s13045-016-0334-6 PMID: 27686492.
- Aguilar Díaz de león, Jesús S., Miyuki Thirumurthy, and Nguyen Ly. "Surface plasmon resonance microscopy identifies glycan heterogeneity in pancreatic cancer cells that influences mucin-4 binding interactions." Plos one 19.5 (2024): e0304154.
