Surface Plasmon Resonance (SPR) can be applied as a convenient, sensitive and label-free technique to study various surface phenomena. One such example is the Vroman effect exhibited by protein adsorption onto surfaces. This important effect arises from the fact that protein adsorption capability onto a surface depends on motility, which is intimately related to its molecular weight. In general, a high molecular weight (HMW) protein adsorbs more strongly than a low molecular weight (LMW) protein. As a result, when a surface is exposed to a solution containing different proteins, it will be covered with more HMW proteins than the LMW ones. This competitive nature of protein adsorption may be used as a mechanism for selective detection and separation of proteins.
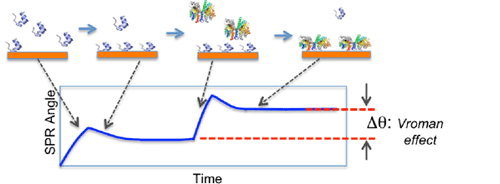
FIG. 1 Schematic illustration of the Vroman effect. A LMW protein adsorbs onto a surface first. Exposing the surface to a HMW protein, the LMW protein is displaced by the HMW protein.
Chae et al. [1] studied proteins with various molecular weights, including aprotinin (0.65 kDa), lysozyme (14.7 kDa), streptavidin (53 kDa), and isolectin (114 kDa) on three different surfaces, including a bare-gold surface and gold surfaces premodified with OH¯- and COOH¯-terminated self-assembled monolayers (SAM). The real-time adsorption and displacement of the proteins were monitored and quantified in real time by SPR. They were able to distinguish these proteins easily based on the Vroman effect.
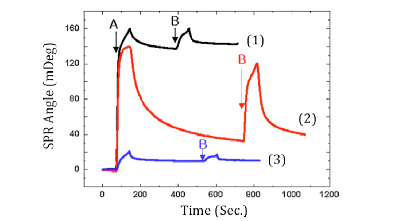
FIG. 2 Real-time SPR displacement pro?les of aprotinin (marked by “A”) by lysozyme (marked by “B”) on (1) the bare-gold, (2) COOH-SAM and (3) OH-SAM surfaces. Group A is selected as LMW proteins and Group B as HWM proteins. LMW proteins are first injected, and adsorption of the proteins on the surfaces is monitored with SPR in real time. HMW proteins are then injected to compete for the surface with the LMW proteins.
In one set of experiment, they started with a surface initially covered with a monolayer of LMW proteins, and studied the displacement of the LMW proteins by HMW proteins. The monolayer formation of the LMW proteins, and subsequent displacement process were monitored in real time and quantified by SPR. FIG. 2 shows an example of the adsorption and displacement profiles of sequentially injected proteins with low and then high molecular weight proteins on bare-gold (black line), COOH¯-terminated SAM (red line) and OH¯-terminated SAM (blue line) surfaces. The first step is to inject 0.05% (W/V) LMW proteins (aprotinin) into the sample cell. The SPR angle increases rapidly, then slows down and approaches saturation. A buffer PBS solution containing no proteins is then injected to wash off weakly bound proteins from the surface, which is observed as a gradual decrease in the SPR angle. The SPR angle does not return to the original value, indicating irreversibly adsorption of the LMW proteins. From the SPR angular shift, the amount of the irreversibly adsorbed LMW proteins is determined. Finally, 0.05% (W/V) HMW proteins (lysozyme) is injected which also leads to an initial rapid increase in the SPR angle. After washing off the weakly bound proteins, the final SPR angle is higher than the angle for the LMW protein monolayer. The difference is the due to the displacement of the LMW proteins by the HMW proteins. Chae at al. also carried out another experiment by reversing the order, namely, starting with HMW proteins on the surface, and following with LMW proteins. The excellent correlation between the SPR results and the Vroman effect suggests that the hydrophobic core of a protein, whose size is proportional to its molecular weight, largely governs its interaction with a solid surface.
Author: Nguyen Ly | Biosensing Instrument | Published January 11, 2016
Acknowledgement: We would like to thank Dr. Chae for sharing his results with us.
DOWNLOAD PDF
Download a PDF of Application Note: 110 – Studying Protein Adsorption Properties with SPR
- Choi, S.; Yang, Y.; and Chae, J. Biosensors & Bioelectronics, 2008, 24: 893-899
