Transporters are large proteins (40–200 kDa) located in the plasma membrane of cells and organelles. They normally span the membrane many times and modulate the transfer of xenobiotics (including nutrients, micronutrients and pharmaceuticals), and endogenous substances such as neurotransmitters, hormones, signaling molecules, vitamins across cellular membranes, tissues or organ barriers.[1] Transporters are clinically important in drug absorption, distribution, and elimination, however, they are notoriously difficult to study. Owing to their hydrophobicity, they are defiant to direct manipulation and can only be removed from the membrane in the presence of detergent to maintain their solubility. Such difficulties help to explain why, to date, fewer than 300 unique membrane protein structures have been resolved.[2]
Radioligand method has traditionally been used for membrane protein binding but there are limitations with the radioactive isotope labeling requirement. Surface Plasmon Resonance (SPR) has been the go-to technique for measuring binding affinity and kinetics in a label free manner. However, measuring membrane proteins binding with the SPR has been a challenge because of the need to immobilize purified receptors on the sensor chip surface. SPR microscopy (SPRm) is a label free technology utilized to study binding interactions of membrane proteins without extracting the proteins from the cell, ensuring intact native conformations of the membrane proteins.[3] Therefore, this technology provides a great value to the research of characterizing binding properties of membrane targets, in particular to membrane transporters, since these proteins are studied in its natural environment.
SPRm 200 system was used to measure the binding of a small molecule (~ 400 Da) to an undisclosed membrane transporter. Two HEK 293 cells were prepared on the SPRm chip: one chip, overexpressed with the target transporter and another chip was prepared as the negative control. Running buffer for both experiments was PBS pH 7.2 0.1% BSA, 100 uM CoCl2. Dilution series of 200 nM, 1/3 dilution, 8 points of the small molecule were injected into each chip.
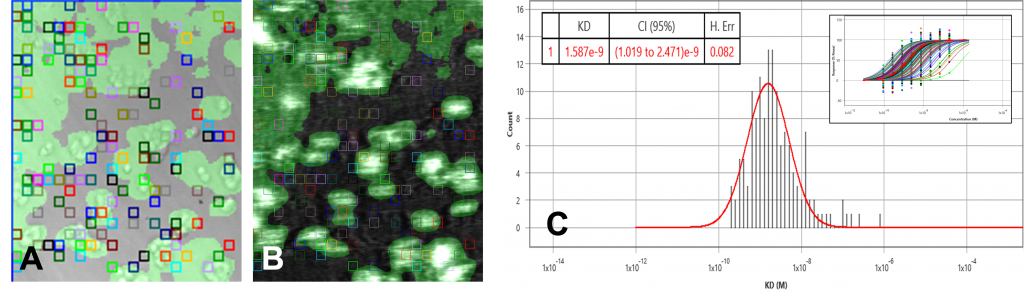
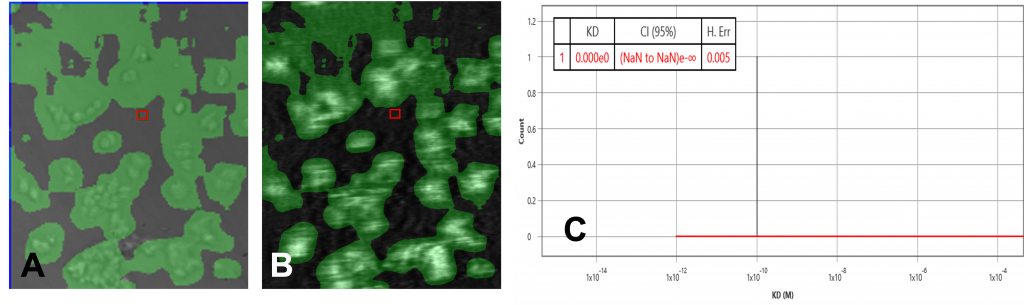
The data set of both overexpressed HEK and negative control runs were analyzed using the ImageSPRTM software. The bright field/SPR images of the positive and negative chip, as well as the KD histograms, presented very different profiles. The overexpressed HEK chip showed binding activities in many regions where the cells are present (Fig 1A, 1B) while the negative chip only showed one binding activity region (Fig 2A, 2B). This binding region in the negative control is likely due to secondary effects such as non-specific binding or target heterogeneity.
The equilibrium analysis KD of the overexpressed cell chip measured 1.57 nM, with 95% confidence interval 1.48 – 1.65. Kon and koff from the kinetic model measured 5 x 106 M-1 s-1 and 4.9 x 10-3 s-1 respectively. The negative control chip histogram showed no significant binding.
In conclusion, SPR microscopy is a powerful tool that measures binding affinity and kinetics of membrane targets that are difficult to measure with traditional technologies.
We thank Dr. Noritaka Furuya and Dr. Kazumasa Yokoyama from Kissei Pharmaceutical Inc for the samples and useful discussions.
DOWNLOAD PDF
Download a PDF of Application Note 142: Small Molecule Binding to Membrane Transporter Using SPRm200
- Keogh, J et al., Membrane Transporters: Fundamentals, Function and Their Role in ADME , Drug Transporters, Volume 1, 2016, pp. 1-56. DOI: 10.1039/9781782623793-00001
- Henderson P.J.F, Comprehensive Biophysics edited by Edward H. Egelman, Volume 8, 2012, pp 265-288, Elsevier B.V. DOI https://doi.org/10.1016/B978-0-12-374920-8.00822-5
- Wang, W et. al., Nature Chemistry 4, 846–853 (2012)
