SPR Microscopy (SPRM) is a powerful tool for measuring the binding kinetics of ligands or drug compounds with membrane proteins, including GPCRs. It has many other unique features that lead to new applications. A recent example is the integration of SPRM with electrochemical methods for molecular binding kinetics quantification.
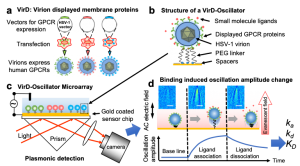
Researchers at Arizona State University and Johns Hopkins University developed a virion oscillator technology using SPRM Impedance. The technology displayed (VirD) GPCRs in the herpesvirus envelope to detect ligand binding kinetics with the GPCRs with a self-assembled virion-oscillator microarray detection (Fig. 1). To measure ligand binding to the GPCRs in the virial envelopes, each oscillator consists of a VirD tethered to a SPR sensor surface with a flexible molecular linker. When applying an alternating electric field normal to the sensor surface with a three-electrode electrochemical setup, where WE, RE and CE are the working (the Au chip), quasi-reference (Ag wire) and counter electrodes (Pt coil), respectively. The virion particle is forced to oscillate with the electric field, and the oscillation amplitude is measured with a SPRM with sub-nm resolution. Upon ligand binding to the GPCRs, the oscillation amplitude changes. Therefore, by measuring the oscillation amplitude in real time with SPRM, the kinetics of ligand binding to the GPCRs is obtained.
FIG. 1 Measuring ligand binding to GPCRs in their native membrane environment with SPRM. a) Herpesvirus (HSV-1) particles are transfected to display human GPCRS (VirD); b) the particles (virions) are tethered to a gold sensing surface via flexible PEG linkers to form virion oscillators; c) VirD-Oscillator microarray are driven by an alternating electrical field into oscillate, and the oscillation amplitudes are detected by a plasmonic imaging technique with sub-nm precision; d) molecular binding events change the oscillation amplitudes, allowing determination of the binding kinetics, described by association (ka) and dissociation (kd) rate constants, and affinity (KD) with SPRM Impedance. Each virion appears as parabolic pattern formed by the scattering a propagating plasmonic wave by the virion.
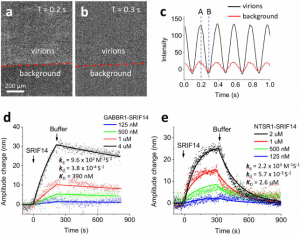
Using SPRM Impedance, the ASU-JHU team measured the oscillation amplitude of the virions on the SPR sensor surface from the average plasmonic imaging intensity and determined the binding kinetic constants. Fig. 2 shows an example of the binding kinetics measurement between peptide ligands and different GPCRs. The measured binding kinetics are distinctively different between intended and off-target ligands. The work demonstrated that the SPRM Impedance system can provide binding kinetics constants via monitoring the plasmonic imaging intensity.
FIG. 2 (a) and (b) VirD oscillators spotted on a SPR sensor chip and imaged with SPRM. Although the individual VirD oscilltors cannot be resolved, the image intensity of each spot oscillates with the applied AC electrical field (c), providing the average oscillation amplitude of the VirD oscillators. The red line in (c) is the image intensity from the background regions, which was subtracted out in data analysis. (d) and (e) show the kinetics of a peptide ligand (SPRIF14 MW=1560) binding to two different types of GPCRs (GABBR1 and NTSR1), where the solid lines are global fitting of the data to the first order kinetics. The extracted kinetic constants are shown on the graphs. Applied voltage: 0.3 V amplitude at 5 Hz. Buffer: PBS.
Author: Nguyen Ly | Biosensing Instrument | Published May 4, 2020
DOWNLOAD PDF
Download a PDF of Application Note 130: Measuring molecular binding kinetics with SPRM Impedance
- G.Z. Ma et al, Journal of the American Chemical Society, 2018 Sep 12;140(36):11495-11501