Understanding conformational changes in proteins and other biomolecules is important because of their critical roles in many biological functions and processes. These conformational changes are often small and fast, making it difficult to measure and monitor. Surface Plasmon Resonance (SPR) is capable to detect rather small conformational changes in surface bound molecules [1]. However, most SPR setups are not suitable for monitoring fast kinetics. BI-SPR 1000 can follow fast kinetics of various processes taking place on surfaces, including conformational changes in proteins (FIG. 1). One such example is the detection of fast redox-induced conformational change in cytochrome c (Cytc) [2].
FIG. 1 Redox reaction of a protein is often accompanied by a conformational change. SPR allows one to detect repaid occurring conformational changes in molecules.
Cytc. was immobilized onto a 3-mercaptopropionic acid (MPA) coated-gold film. Initially, the potential of the gold film was held at +0.2 V (v.s. Ag/AgCl) with a potentiostat and the protein was in the oxidized state. By stepping the potential to –0.1 V, the protein was switched to the reduced state. After holding it in reduced state for 2.5 sec., the protein was re-oxidized by stepping the potential back to +0.2 V. The SPR angle shift as a function of time was recorded during the reduction and the oxidation processes. In the absence of the proteins on the gold film, stepping the potential also causes shifts in the SPR angle, due to change of charge density in the film, but the rise and decay is as fast as a few ms (FIG. 2a). These finite rise and decay time constants are determined by the response of the electrochemical cell.
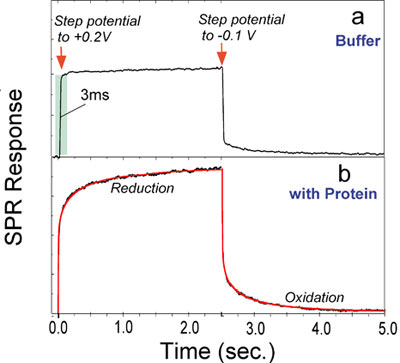
FIG. 2 Kinetics of redox-induced conformational changes in cytochrome c immobilized on a 3-mercaptopropionic acid coated-gold sensor chip. (a) In the absence of the protein, stepping the potential between -0.1 V and +0.2 V causes rapid changes (~3 ms) in the SPR angle, due to charging of the sensor chip. (b) In the presence of the protein, stepping the potential switches the protein between the oxidized and reduced states, and the associated conformational change in the protein is monitored by SPR.
As shown in FIG. 2, with Cytc on the surface, a much greater and slower SPR angular shift associated with the reduction and the oxidation was observed (FIG. 2b). The kinetics of the SPR data cannot be with simple exponential functions. However, excellent fits were obtained by using a stretched exponential function, exp[-(t/t)b], with b~ 0.5. Stretched exponential processes are quite common in complex systems, such as proteins. A well-studied example is the binding of CO or O2 to myoglobin [3-8]. Although the same explanation may apply to the electron transfer reaction-induced conformational changes in Cytc, more experiments will be needed to pin down the origin of the stretched exponential conformational change.
The time constants determined by fitting the experimental data with the stretched exponential function are 210 and 140 ms for the oxidation and the reduction, respectively, which are 2-3 orders of magnitude slower than the electron transfer time constant. Therefore, the conformational change lasts long after the electron transfer is over. This observation indicates that the electron transfer reaction-induced conformational changes in Cytc involve a slow collective structural relaxation rather than simply a local change in the axial ligands. The conformational changes following an oxidation are slower than those following the reduction of Cytc, which agrees with an earlier circular dichroism study.
Author: Nguyen Ly | Biosensing Instrument | Published January 1, 2008
DOWNLOAD PDF
Download a PDF of Application Note 101: Fast Kinetics of Conformational Changes in Proteins
- S. Boussaad, J. Pean, and N. J. Tao, Anal. Chem., 72:222 (2000).
- S. Boussaad and N.J. Tao J. Electroanal. Chem., 554-555, 233-239 (2003).
- C. K. Chan, J. Hofrichter, and A. W. Eaton, Science 274:628.
- W. Colon, A. G. Elove, L. P. Wakem, F. Sherman, and H. Roder, Biochemistry 35:5538 (1996).
- A. M. Berghuis and G. D. Brayer, J. Mol. Biol. 223:959 (1992).
- T. Takano and R. E. Dickerson, J. Mol. Biol. 153:79 (1981).
- H. Sota, Y. Hasegawa, and M. Iwakura, Anal. Chem. 70:2019 (1998).
- R. H. Austin, K. W. Beeson, L. Eisenstein, and H. Frauenfelder, Biochem. 13:5355 (1975).
